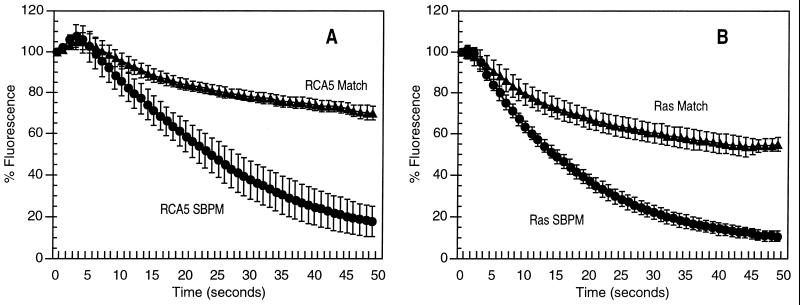
Figure 4.
Discrimination of match from SBPM for genomically different oligonucleotide hybrids on the same chip. ATA5 match and SBPM capture oligonucleotides were addressed to all five electrodes of C1 and C2, respectively. Ras match and SBPM capture oligonucleotides were directed to C4 and C5, respectively. RCA5 and Ras reporter oligonucleotides were passively hybridized to the previously addressed capture oligonucleotides. The hybridization conditions were 1 μM for each oligonucleotide in 50 mM sodium phosphate/500 mM NaCl, pH 7.4, in 15 μl at room temperature for 5 min. Electric field denaturations were done after equilibration in 20 mM sodium phosphate buffer, as described in the legend of Fig. 3 and below. Images were captured at 1-sec intervals, and average pixel intensity (API) was determined over the electrode area. Data were normalized by dividing the API at each time point by the API at t = 0, × 100. Points represent the average percent fluorescence remaining on 15 replicate test sites, from three separate chips in three separate experiments. Error bars represent the standard deviation at each point. The electric field sweeps took approximately 45 sec. (A) Test sites containing either RCA5 Match or RCA5 SBPM sequence (19-mers) were subjected to current pulses of 0.6 μA, 0.1 sec on, 0.2 sec off, 150 cycles. (B) Test sites containing either Ras Match or Ras SBPM (22-mers) received current pulses of 1.5 μA, 0.1 sec on, 0.2 sec off, 150 cycles.