Fig. 1.
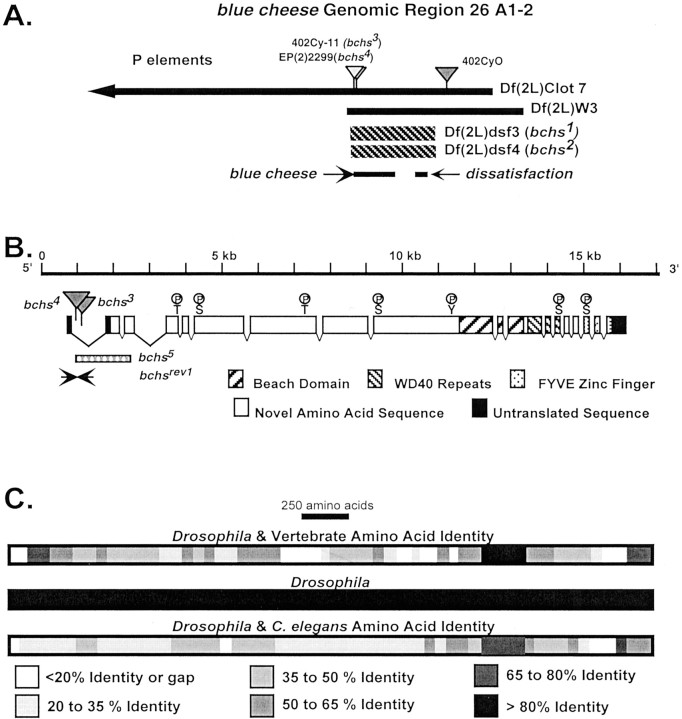
The bchs region, mutations, protein structure, and species alignment. A, The location and extent of deletions uncovering bchs are illustrated. The map of the bchs genomic region and deletion boundaries is as described previously [Finley et al. (1998); see flybase website in Materials and Methods]. The location and arrangement of thebchs and dsf genes are indicated, as are the 402Cy P-element (w+;darkgray) and P-elements generatingbchs mutations (bchs3and bchs4; light gray). The 402Cy P-element is the parent of the Df(2L)dsf3 and Df(2L)dsf4 deletions and the bchs3 insertion. It is carried on the CyO chromosome used as a wild-type control in aging experiments. B, The intron/exon structure ofbchs was determined by comparison and alignment of cDNA and genomic sequences. The locations of BEACH, WD40, and FYVE protein motifs and potential phosphorylation sites are indicated. Sites of P-element insertion are indicated. The extent of thebchs5 internal deletion (determined by DNA sequencing) is indicated by the hatched bar. A precise excision derivative, thebchsrev1 allele, was determined by DNA sequencing and is indicated by arrows. C,Drosophila, vertebrate, and nematode BCHS protein alignments were generated by comparison of the inferred amino acid sequences. The overall size, protein domains, and amino acid sequence of the different BCHS proteins are conserved, including the first 2500 amino acids possibly identifying functional motifs within this region. The identified domains within the C-terminal region have the highest amino acid identity over divergent phyla, reaching >80% over extended regions of the protein.