Abstract
The transcriptional and chromatin profile of the promoter, first exon and first intron of the human TH gene were analyzed in human neuroblastoma BE(2)-C-16 and human renal carcinoma 293FT cell lines. The latter is a cell culture system that is not permissive for TH gene expression, whereas the former has a 50% cell fraction that tests positive for TH. The engineering of a 6.3 kb recombinant human TH promoter revealed the presence of repressors of transcription between positions (−6,244/−194). The addition of a 1.2 kb fragment of the first intron of the human TH gene (+730/+1,653) enhanced transcriptional activity of the recombinant promoter. However, both constructs were not specific for TH-positive BE(2)-C-16 cells. Chromatin immunoprecipitation (Chip) analysis was carried out on BE(2)-C-16 and 293FT cells to probe sequences of promoter, first exon and first intron of the human TH gene from position (−448/+1,204). The presence of nucleosomes was observed approximately from position (−20/+473) in both cell lines. Chip analysis was then conducted to determine the acetylation of various lysine residues of H3 and H4 in both cell lines. All analyzed lysine residues of H3 and H4 were acetylated in BE(2)-C-16 cells, whereas 293FT cells tested positive for acetylation only in the external lysine residues of the histone tail. Our data are compatible with an active TH gene expression in a 50% cell fraction of BE(2)-C-16 cells. Further analysis of epigenetic programming might lead to the identification of the factors that determine TH gene expression specifically in dopaminergic neurons.
The elucidation of the mechanism of human tyrosine hydroxylase (TH) gene regulation is one of the key issues in the field of neurology. The function of TH consists of catalyzing tyrosine hydroxylation in the synthesis of L-dopa (Nagatsu et al., 1964), which is the rate-limiting step in the generation of catecholamine neurotransmitters of the central and peripheral nervous systems (Zigmond et al., 1989). There are a number of important reasons to continue to explore the complex mechanism of TH gene regulation. For instance, aberrant TH gene expression is observed in alcoholism and in psychiatric illnesses, such as schizophrenia and bipolar disorder (Ishiguro et al., 1998). In addition, the degeneration of TH-positive dopaminergic neurons of the substantia nigra is associated with Parkinson's disease (Moore, 2003).
Many studies have been conducted to characterize the factors that come into play in TH gene regulation. These studies focused mainly on the human (Kessler et al., 2003; Romano et al., 2005; Jin et al., 2006), mouse (Kim et al., 2003a) and rat models (Gandelman et al., 1990; Kim et al., 2003b). In a previous report, a 13 kb DNA fragment containing the human TH promoter was isolated from a genomic DNA library, sequenced and utilized to generate both a transgenic animal model (Kessler et al., 2003) and a variety of recombinant minimal TH promoters assembled in a self-inactivating lentiviral vector system (Romano et al., 2005). All the recombinant human TH promoters were engineered to drive the expression of green fluorescent protein (GFP). Although high levels of GFP expression were readily observed in TH-positive cells of the substantia nigra of embryonic and adult transgenic mice, subsequent studies revealed remarkable differences in TH gene regulation between the human and mouse models. Comparative analysis of the sequences of the human, mouse and rat TH promoters revealed only five small evolutionary conserved regions (CRs) of high homology (Kessler et al., 2003). The degree of homology between the human and mouse TH promoters is about 46.6% (determined with a Clustalx program; Romano et al., 2005), whereas the human and rat TH promoters share only a 30% degree of homology (Gandelman et al., 1990; Kim et al., 2003b). The five CRs were placed upstream of the first–194 bp from the transcription start of the human TH promoter and the first 35 bp of the untranslated messenger RNA leader of the human TH gene (Romano et al., 2005). This human TH minimal promoter was linked to GFP and assembled in a self-inactivating lentiviral vector system for the in vitro transduction of human neuronal progenitor cells (hNPCs) and mouse primary striatal and Substantia nigra cells (Romano et al., 2005). Transduced cells were then treated in vitro with a mixture of differentiating agents to enhance TH expression (Du and Iacovitti, 1997). Interestingly, the human TH minimal promoter encoding for the five CRs exhibited a significant degree of specific gene expression only in induced TH-positive hNPCs (Romano et al., 2005), while it failed to do so in TH-positive differentiated mouse primary striatal cells and in differentiated mouse substantia nigra cells (Romano et al., 2005). This finding is consistent with differences in the mechanism of TH gene regulation between the human and mouse systems. Another striking difference between the human and murine models was observed in a more recent study on the nuclear orphan receptor NR4A2 (Nurr1), which did not affect human TH gene expression in hNPCs, in contrast to the mouse and rat systems (Jin et al., 2006). Nurr1 is required to transactivate mouse TH minimal promoters (Iwawaki et al., 2000; Kim et al., 2003a), as well as other unrelated promoters, such as the osteocalcin promoter in mouse osteoblasts (Pirih et al., 2004). However, TH gene expression did not depend on a direct Nurr1-mediated transactivation in the human model (Jin et al., 2006).
Indeed, TH gene regulation is achieved through different mechanisms among species, as indicated also by a study conducted on three scaffold/matrix attachment regions (S/MAR) present in the promoter at positions (−186/−16) and in the first intron (+645/+755 and +835/+945) of the human TH gene (Lenartowski et al., 2003). The association between nuclear matrix of certain cell types and various DNA binding proteins may constitute a subcellular structure in chromatin organization, which, in turn, may result in a particular tissue-specific gene regulation (van Wijnen et al., 1993). A comparative analysis was carried out between human and bovine TH genes in binding nuclear matrix obtained from bovine brain and liver tissues (Lenartowski et al., 2003). In contrast to the human TH gene (Lenartowski and Goc, 2002), the association between bovine TH gene and the nuclear matrix was not tissue specific, although the position of the matrix binding region is conserved in both systems (Lenartowski et al., 2003). The aim of this study was to characterize the transcriptional and chromatin modification profile of the promoter, first exon and first intron of the human TH gene, in order to better understand the mechanism of TH gene regulation in human cell culture models. Our findings indicate that chromatin remodeling might play a relevant role in conferring tissue specific gene expression of the human TH gene.
Materials and Methods
Plasmids
Plasmid pGR401 contains a 6.3 kb human minimal TH promoter driving GFP (−6,244/+35) and assembled in a self-inactivating lentiviral vector system (Lois et al., 2002). The 6.3 kb DNA fragment was excised by a partial BamHI digestion from plasmid pMAK288 (Kessler et al., 2003) and ligated into the dephosphorylated BamHI restriction site of a plasmid based on the pFUGV self-inactivating lentiviral vector system (Lois et al., 2002; Romano et al., 2005).
Plasmid pGR435 contains a 6.3 kb human minimal TH promoter driving the expression of GFP. This plasmid derives from plasmid pGR401 and is no longer a self-inactivating lentiviral vector system, as the 5′ long-terminal repeat (LTR) of HIV-1 was deleted along with the enhancer of the human cytomegalovirus (hCMV) by MluI and PacI digestion. The plasmid was then filled in with Klenow fragment and ligated to generate plasmid pGR435.
Plasmid pGR436 contains a 6.3 kb human TH minimal promoter driving the expression of GFP (−6,244/+35) and a 1.2 kb fragment of the first intron of the human TH gene from position (+730/+1,653) (O'Malley et al., 1987). The 1.2 kb fragment of the first intron of the human TH gene was obtained via PCR and placed downstream of the GFP cassette of plasmid pGR435, therefore, also plasmid pGR436 is not a self-inactivating lentiviral vector system. The template for PCR was genomic DNA derived from human BE(2)-C neuroblastoma cell line. The sequence of the sense primer was: 5′-gaattcTGGAGGCCTGGGCACAGCGGCATCCCCTGT-3′ (positions +730/+758). Lower case letters indicate the EcoRI restriction site contained in the overhang of the sense primer. The sequence of the antisense primer was: 5′-atcgatCATGAAGGGGAGCAGCAGAAGCCCCTCCCA-3′ (positions +1,623/+1,652). Lower case letters indicate the ClaI restriction site contained in the overhang of the antisense primer. The PCR conditions were as follows: an initial denaturation step of the template at 94°C for 1 min, followed by 1 min interval at 48°C to allow for the annealing of the primers to the template and 5 min incubation at 72°C for polymerase elongation of the primers. This cycle was repeated 35 times. At the end of the 35 cycles, an additional incubation at 72°C for 20 min was included to allow for the completion of the amplification. The PCR product was isolated and ligated into EcoRI and ClaI digested and dephosphorylated pGR435. The inserted PCR fragment was then sequenced to check for possible misincorporations.
Cell lines
Human neuroblastoma BE(2)-C cell line was purchased from ATCC and grown in DMEM/F12 (Invitrogen, Carlsbad, CA) supplemented with 10% (v/v) FBS (Invitrogen), 0.5 mM sodium pyruvate (Invitrogen), 0.05 mM non-essential amino acids (Invitrogen) and 2 mM l-glutamine (Invitrogen). Human neuroblastoma BE(2)-C cell line was subcloned by limiting dilution to generate BE(2)-C-16. TH is expressed in a 20% fraction of the parental BE(2)-C cell line, whereas about a 50% fraction of the subclone BE(2)-C-16 tests positive for TH.
Fetal human renal carcinoma cell line 293FT was purchased from Invitrogen and was grown in Dulbecco's minimum essential medium (DMEM) (Invitrogen) supplemented with 10% (v/v) heat inactivated FBS (Invitrogen) and 2 mM l-glutamine (Invitrogen).
Human breast carcinoma MCF-7 cell line was purchased from ATCC and grown in DMEM (Invitrogen) supplemented with 10% (v/v) FBS (Invitrogen) and 2 mM l-glutamine (Invitrogen).
Lentiviral-mediated gene transfer
Lentiviral vector stocks were generated transiently for the gene transduction of target cells as described elsewhere (Soneoka et al., 1995; Lois et al., 2002). Transient calcium phosphate DNA transfection of 293FT cell line was carried out as described (Pear et al., 1993; Soneoka et al., 1995). Lentiviral vector stocks were harvested and used for the transduction of target cells as described elsewhere (Soneoka et al., 1995; Lois et al., 2002).
Transfection experiments of BE(2)-C-16 and 293FT cell lines
Plasmids pGR435 and pGR436 were transfected into BE(2)-C-16 and 293FT cells by using FuGENE 6 Transfection Reagent (Roche, Indianapolis, IN) following manufacturer's instructions.
Antibodies and immunofluorescence
Rabbit antibodies to tyrosine hydroxylase (TH) were purchased from Pel-freez Biological (Rogers, AK) and used at a dilution 1:100 (v/v) in phosphate buffered saline (PBS). Rhodamine-conjugated secondary antibody to rabbit immunoglobulins was purchased from Jackson ImmunoResearch (West Grove, PA) and used at a dilution 1:50 (v/v) in PBS.
Cells were fixed with 4% (w/v) paraformaldehyde in PBS at room temperature for 10 min. Immunofluorescence assay was carried out as described elsewhere (Du and Iacovitti, 1997; Kessler et al., 2003). Endogenous TH expression was detected with rhodamine-conjugated antibodies. Slides were analyzed on a Nikon-Scanalytic Image System as described elsewhere (Yang et al., 2004). Numbers of green, red and overlapped green and red cells were counted manually. Three different fields, at a magnification of 20×, were taken per each well in order to score green, red and yellow cells per each sample.
Antibodies for Chip analysis were purchased from Upstate (Lake Placid, NY). Chip analysis was carried out with antibodies to unmodified H3, Ac-K9/18 H3, Ac-K14 H3, Ac-K23 H3, Ac-K27 H3, pan antibodies to H4, Ac-K5 H4, Ac-K8 H4, Ac-K12 H4, Ac-K16 H4, and penta Ac-(K5, 8, 12, 16, 20) H4. Immunoprecipitations were carried out with 1 μg of antibody per sample, as described in the Chip analysis section.
Chip analysis
Chip analysis was carried out with a kit purchased from Upstate following manufacturer's instructions. Briefly, BE(2)-C16 and 293FT cell lines were grown in 10 cm tissue culture dishes until 80% confluence. Cells were then fixed with 1% formaldehyde (Sigma, St. Louis, MO) for 10 min at 37°C. At the end of the incubation, cells were washed twice of ice-cold PBS, scraped off the tissue culture dishes in 10 ml of ice-cold PBS, spun down and lysed on ice for 10 min with 200 μl of lysis buffer and protease inhibitors (Roche). Cell lysates were then sonicated on ice three times with a Fisher Model 500 Sonic Dismembrator (Fisher, Pittsburgh, PA), at maximum amplitude (70%), for 10 sec with 15 sec intervals. Immunoprecipations were carried out with 1 μg of antibody per each sample, as described by the manufacturer (Upstate). Immunoprecipitates were then washed, eluted out of the beads, extracted with phenol–chloroform 1:1 (v/v) (Sigma), ethanol precipitated, washed with 70% ethanol, air dried, resuspended with 30 μl of nuclease-free distilled water (Ambion, Austin, TX), an aliquot of 3 μl was then used for PCR analysis, while the remaining part was frozen at −20°C. Five sets of sense and antisense primers were designed for Chip analysis of the promoter, first exon and first intron of the human TH gene from position −448 to +1,204. The sequences and coordinate of the primers are listed in Table 1. The PCR conditions were as follows: an initial denaturation step of the template at 94°C for 1 min, followed by 1 min interval at 48°C to allow for the annealing of the primers to the template and 1 min incubation at 72°C for polymerase elongation of the primers. This cycle was repeated 35 times. An additional incubation at 72°C for 20 min was included to allow for the completion of the amplification. An aliquot of 10 μl was run on a 1% agarose gel to visualize and photograph PCR products.
TABLE 1.
Coordinates and sequences of the five sets of primers for Chip analysis of the promoter, first exon and first intron of the human TH gene
| Set number, coordinates and type of primer | Sequence (from 5′ to 3′) |
|---|---|
| First set, (−448; −418), sense primer | GTCTACGAGACACACGGCCTGGAATCTTCT |
| First set, (−50; −20), antisense primer | CAAAGCCCCCTCTGGGTCCCCCACCTTCCC |
| Second set, (−87; −57), sense primer | CATCAGGCACAGCAGGCAGGGGTGGGGGAT |
| Second set, (+166; +196), antisense primer | CCCTGGGCTCCGGTCCACTGCGGCCGCCGG |
| Third set, (+106; +136), sense primer | GTCTGAGCTGGACGCCAAGCAGGCAGAGGC |
| Third set, (+512; +542), antisense primer | CAGAGGGAGGCAGGGGCAGGTGGGAGTAGG |
| Fourth set, (+443; +473), sense primer | AGATGGCCAGAGGTCCCCGGCTGCTGCACC |
| Fourth set, (+872; +902), antisense primer | GAGCCCAAGGTTCTGAGTGCCCAAGGAGGC |
| Fifth set, (+813; +843), sense primer | GCTTTTCAGCCCACAGGAGGGGTCTTCGGT |
| Fifth set, (+1174; +1204), antisense primer | CATGAAGGGGAGCAGCAGAAGCCCCTCCCA |
Chip analysis was also conducted on sonicated genomic DNA from 1% formaldehyde fixed human MCF-7 cell line, in order to have a positive control for the antibody to unmodified H3. Two primers were used to amplify a 283 bp sequence of the endogenous estrogen receptor-α as described elsewhere (Macaluso et al., 2003, 2006).
Results
Profile of gene expression and specificity for human TH-positive cells of the 6.3 kb minimal promoter and 1.2 kb fragment of the first intron of the human TH gene
A previous study showed that a human TH minimal promoter driving GFP termed pGR382 (−194/+35) had a considerably high transcriptional activity in hNPCs, while it was silent in human skin carcinoma A431 cell line (Romano et al., 2005). However, this construct was not specific for TH-expressing differentiated hNPCs (Romano et al., 2005). Two new constructs were generated for a twofold purpose: (I) to determine whether genetic elements of the human TH promoter comprised in the region (−6,244/−194) might improve specific gene expression for TH-positive human neuroblastoma BE(2)-C-16 cell line and (II) to study the function of a 1.2 kb first intron fragment of the human TH gene encoded in the region (+730/+1,653) in terms of transcriptional activity and specificity for TH-positive BE(2)-C-16 cells (Fig. 1). The splicing signals were removed from the intron fragment (+730/+1,653), which contains the microsatellite repeat HUMTH01 and the S/MAR at position (+835/+945). Plasmid pGR435 contains the 6.3 kb human TH minimal promoter driving GFP expression (Fig. 1). The intron fragment was placed downstream of the GFP cassette, to generate plasmid pGR436 (Fig. 1). These two plasmids were used in transient transfection experiments in human neuroblastoma BE(2)-C-16 and human renal carcinoma 293FT cell lines. The transfecting reagent was FuGENE 6 (Materials and Methods). Lentiviral-mediated gene transfer was efficient in expressing GFP driven by the 6.3 kb human TH minimal promoter in transduced human neuroblastoma BE(2)-C-16 cells, albeit at very low levels (data not shown). However, the presence of the 1.2 kb intron fragment proved detrimental for lentiviral-mediated gene transfer, which failed to transduce target cells. For this reason, our study had to rely on conventional plasmid DNA transfection protocols. As mentioned, the 5′-LTR of HIV-1 and the hCMV enhancer were removed from the parental lentiviral vector plasmids to generate plasmids pGR435 and pGR436, in which GFP is under the control of the 6.3 kb human TH minimal promoter in transfected cells (Materials and Methods).
Fig. 1.
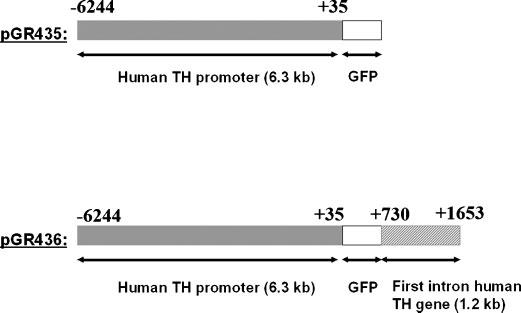
Map of plasmids pGR435 and pGR436. The human TH minimal promoter comprises a sequence from (−6,244/+35), whereas the 1.2 kb fragment of the first intron of the human TH gene contains a sequence from (+730/+1,653). The 1.2 kb first intron fragment was placed downstream of GFP cassette (plasmid pGR436). The coordinates of the promoter and first intron have been reported on top of the bars that depict the two constructs.
Interestingly, plasmid pGR436 exhibited 3.4-fold increase in GFP intensity than plasmid pGR435 (Fig. 2), indicating that the 1.2 kb intron fragment of the human TH gene (+730/+1,653) plays a relevant role in enhancing transcriptional activity of the 6.3 kb human TH minimal promoter. GFP intensity was evaluated in Photoshop. Both plasmids could not express GFP in human 293FT cell line (data not shown).
Fig. 2.
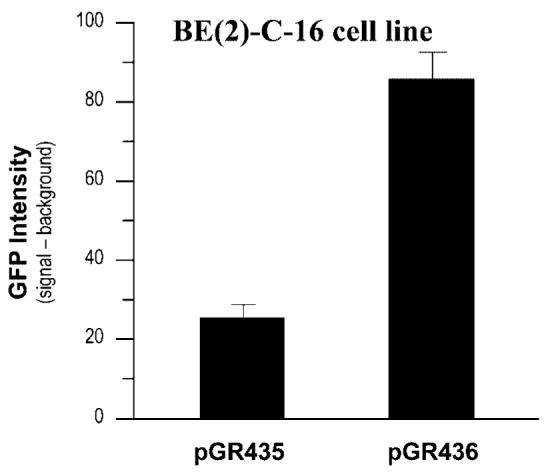
This graph shows the increase in GFP expression levels driven by the 6.3 kb human TH minimal promoter in the presence of the 1.2 kb first intron fragment of the human TH gene (plasmid pGR436) following transfection into human neuroblastoma BE(2)-C-16 cell line. Plasmid pGR435 contains only the 6.3 kb human TH minimal promoter. GFP expression and background were quantified in Photoshop. Each difference (GFP signal–background) represents the mean value +/− standard deviation of three determinants.
The degree of specific gene expression of plasmid pGR436 for TH-positive BE(2)-C-16 cells was determined as described (Romano et al., 2005). Briefly, GFP expression in BE(2)-C-16 transfected cells was monitored under an ultraviolet (UV) microscope. Human endogenous TH was detected immunocytochemically with rhodamine-conjugated antibodies on paraformaldehyde fixed cells (Materials and Methods). Plasmid pGR436 did not show any significant degree of specificity for TH-expressing human neuroblastoma BE(2)-C-16 cells (Fig. 3). A similar finding was observed for the constructs pGR401 and pGR435, which lack the 1.2 kb intron fragment of the human TH gene (data not shown).
Fig. 3.
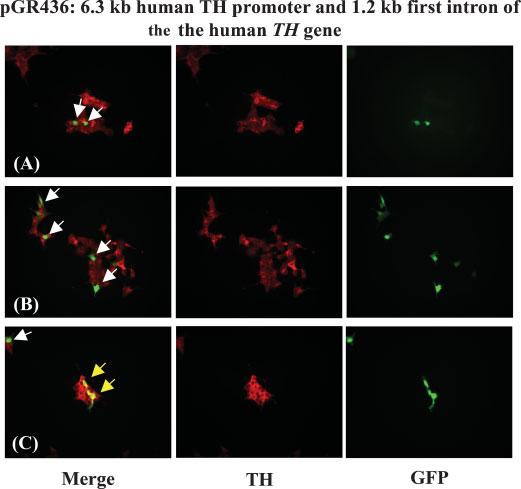
Immunocytochemical localization of TH expression and detection of GFP in human neuroblastoma BE(2)-C-16 cells after transfection with plasmid pGR463 (6.3 kb human TH minimal promoter, GFP and 1.2 kb first intron fragment of the human TH gene). TH was stained with rhodamine. Yellow arrows depict two overlaps in TH and GFP expression in BE(2)-C-16 cells. White arrows depict GFP signals that are not overlapped with TH.
In summary, GFP driven by the 6.3 kb human TH minimal promoter (−6,244/+35) is weakly expressed and is not specific for TH-positive human neuroblastoma BE(2)-C-16 cells. The addition of the 1.2 kb first intron fragment (+730/+1,653) of the human TH gene enhanced GFP expression driven by the 6.3 kb human TH minimal promoter in human neuroblastoma BE(2)-C-16 cells, but without improving specific gene expression for TH-positive cells.
Mapping of chromatin localization in the promoter, first exon and first intron of the human TH gene between positions (−448/+1,204)
Genetic analysis has so far identified elements that are required for efficient TH gene expression in brain derived cells, but without any significant specificity for TH-expressing human neuroblastoma BE(2)-C-16 cells. Indeed, studies on chromatin remodeling might shed useful insights in better understanding the mechanism of human TH gene regulation. To this end, genomic DNA was extracted both from paraformaldehyde fixed human BE(2)-C-16 and human 293FT cell lines and used for qualitative Chip analysis (Materials and Methods). A pan antibody to H4 was utilized to immunoprecipitate chromatin-DNA complexes and a part of five sets of primers was then used for PCR analysis (Table 1). The region under investigation comprised the promoter, first exon and first intron of the human TH gene between positions (−448/+1,204). The five sets of sense and antisense primers were designed to amplify DNA segments in the range of 300–400 bp (Table 1). Each set of sense and antisense primers had some overlapping region with the flanking one (Table 1) and was tested by PCR on genomic DNA extracted from 293FT cell line as positive control (Fig. 4; lower part of the panel). Negative controls for the five sets of PCR primers used in this study were obtained by conducting ChIP without antibody to H4 on paraformaldehyde fixed and sonicated genomic DNA derived from human BE(2)-C-16 and 293FT cell lines (data not shown). Interestingly, Chip analysis revealed the presence of a small cluster of nucleosomes approximately between positions (−20/+473) in both cell types (Fig. 4). These experiments were conducted three times for both cell types.
Fig. 4.
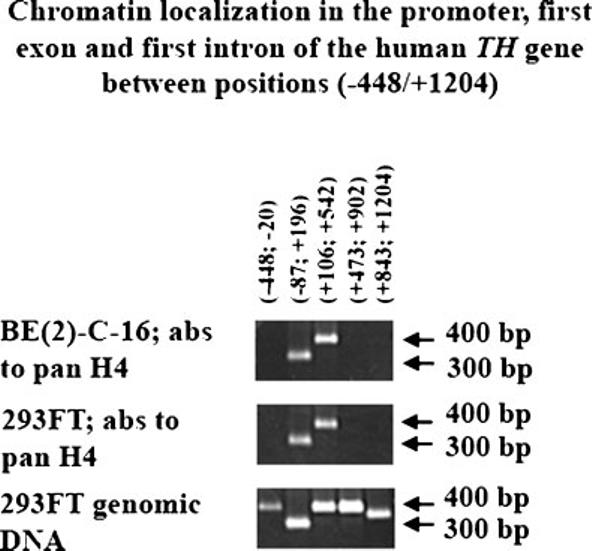
Chip analysis of chromatin localization in the promoter, first exon and first intron of the human TH gene between position (−448/+1,204). Antibodies (abs). Pan antibodies to H4 were used for Chip analysis of genomic DNA and chromatin complexes extracted from human neuroblastoma BE(2)-C-16 and human renal carcinoma 293FT cell lines (Materials and Methods). Five sets of sense and antisense primers were used for PCR analysis of Chip products (Table 1). The coordinates of the genomic DNA regions analyzed are reported on top of the first part. Two arrows depict 300 and 400 bp PCR products per each of the three parts (right hand-side). Chip analysis of genomic DNA and chromatin complexes extracted from human neuroblastoma BE(2)-C-16 cell line is shown in the top part, whereas the middle part shows the Chip analysis products of genomic DNA and chromatin complexes extracted from human renal carcinoma 293FT cell line. The lower part shows PCR products obtained for the five sets of sense and antisense primers using as template genomic DNA extracted from 293FT cell line.
Analysis of H3 and H4 tail acetylation in the small cluster of nucleosomes of the promoter, first exon and first intron of the human TH gene between positions (−20/+473)
After establishing that there was no difference in terms of chromatin localization in the promoter, first exon and first intron of the human TH gene between human neuroblastoma BE(2)-C-16 and human renal carcinoma 293FT cell lines (Fig. 4), we proceeded with the analysis of tail acetylation of H3 and H4 of the nucleosomes detected by Chip analysis of both cell lines between positions (−20/+473). For this study, we used a part of antibodies to unmodified H3, bi-Ac-H3-K9/18, Ac-H3-K14, Ac-H3-K23, Ac-H3-K27, pan antibodies to H4, Ac-H4-K5, Ac-H4-K8, Ac-H4-K12, Ac-H4-K16, and penta-Ac-H4-(K5, 8, 12, 16, 20). Only two sets of primers were used for PCR analysis of regions (−87/+196) and (+106/+542), respectively. All lysine residues of H3 and H4 tested positive for acetylation in BE(2)-C16 cell line (Fig. 5; part A), whereas 293FT cell line tested positive for acetylation of H3-K14, H3-K23, H4-K5 and H4-K8 (Fig. 5; part B). As expected, pan antibodies to H4 tested positive for both cell lines (Fig. 5; parts A and B). Interestingly, the antibody to unmodified H3 tested negative for Chip analysis of human neuroblastoma BE(2)-C-16 and human renal carcinoma 293FT cell lines (Fig. 5; parts A and B). A set of two primers was used to amplify a 283 bp sequence of the human endogenous estrogen receptor-α as positive control for the antibody to unmodified H3 (Macaluso et al., 2003, 2006) (data not shown). Also in this case, negative controls for the two sets of PCR primers used in this experiment were obtained by conducting ChIP analysis without antibodies to histones on paraformaldehyde fixed and soniated genomic DNA derived from human BE(2)-C-16 and 293FT cell lines (data not shown). In summary, Chip analysis revealed a much higher degree of histone tail acetylation in human neuroblastoma BE(2)-C-16 cells than in human renal carcinoma 293FT cells.
Fig. 5.
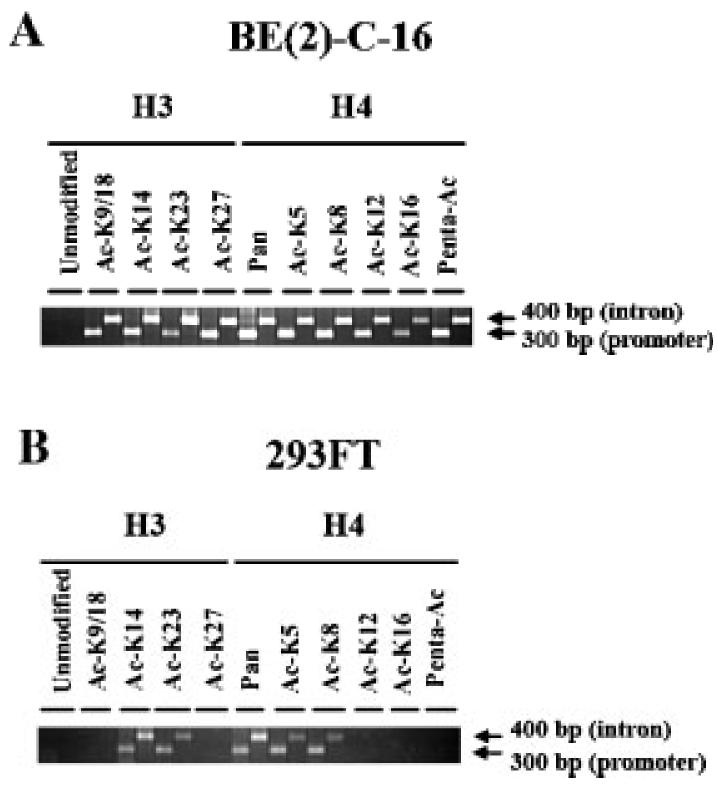
Chip analysis of H3 and H4 tail acetylation in human neuroblastoma BE(2)-C-16 cell line (part A) and human renal carcinoma 293FT cell line (part B). PCR analysis on Chip products was conducted per each antibody with two sets of primers annealing to regions (−87/+196) and (+106/+542) of the promoter, first exon and first intron of the human TH gene in both cell types. Two arrows depict 300 and 400 bp PCR products in parts A and B (right hand-side), which correspond to the PCR signals associated with region (−87/+196) and (+106/+542), respectively. The part of antibodies used for Chip analysis is divided into two groups: H3 and H4 (parts A and B). The specificity of each antibody is reported either below the annotation H3 or H4. The antibody to unmodified H3 is the only one that tested negative for both cell lines. Two primers were used to amplify a 283 bp sequence of the endogenous estrogen receptor-alpha as positive control for the antibody to unmodified H3 (Macaluso et al., 2003, 2006) (data not shown).
Discussion
Our new study focused on two main aspects related to the biology of human TH gene regulation: (I) effects of the 1.2 kb first intron region (+730/+1,653) on the transcriptional activity of a recombinant 6.3 kb human TH minimal promoter, which encoded for the region (−6,244/+35); (II) analysis of the chromatin structure in the region (−448/+1,204) of the promoter, first exon and first intron of the human TH gene in two cell types: human neuroblastoma BE(2)-C-16 and human renal carcinoma 293FT cell lines. The latter one is a cell culture model that is not permissive for TH gene expression, whereas the former cell line has a 50% cell population that is permissive for TH gene expression.
In a previous study, a human TH minimal promoter encoding for the region (−194/+35) exhibited a considerably high transcriptional activity in hNPCs (Romano et al., 2005). However, addition of genetic elements located upstream of the region (−194/+35) reduced transcriptional activity of recombinant human TH minimal promoters, indicating that a number of repressors of transcription are present inside the promoter (Kim et al., 2003b; Romano et al., 2005). Deletion analysis found a repressor of transcription in the region (−1,232/−1,210) of the human TH promoter (Kim et al., 2003b), whereas a study on evolutionary CRs among human, mouse and rat TH promoters identified another sequence encoding for repressor(s) of transcription in the region (−2,423/−2,327), which corresponds to CR-V (Romano et al., 2005). The transcriptional activity of a 6.3 kb human TH minimal promoter encoding for the region (−6,244/+35) was also very low (Fig. 2). However, the addition of the 1.2 kb region (+730/+1,653) of the first intron of the human TH gene (Fig. 1) could enhance the transcriptional activity of the 6.3 kb human TH minimal promoter in human neuroblastoma BE(2)-C-16 cell line (Fig. 2). Thus, genetic elements in the region (−6,244/+35) of the human TH promoter and in the region (+730/+1,653) of the first intron of the human TH gene are required to optimize GFP expression specifically in human neuroblastoma BE(2)-C-16 cell line, while no ectopic GFP expression was ever detected in human renal carcinoma 293FT cell line. However, the expression of the transgene was not specific for TH-positive BE(2)-C-16 cells, indicating that further studies are indeed required to characterize the regulatory mechanism of human TH gene expression specifically in dopaminergic neurons such as those of the substantia nigra. Regardless of the great deal of effort expended by many investigators in analyzing various genetic elements of the promoter, first exon and first intron of the human TH gene, the mechanism of TH gene regulation remains elusive. Some studies shed useful insights in clarifying a confusing issue related to the comparison of TH gene regulation among various species, which is achieved through different mechanisms (Lenartowski and Goc, 2002; Lenartowski et al., 2003; Romano et al., 2005; Jin et al., 2006; Kelly et al., 2006). In this respect, a first important observation derived from the comparative analysis between the sequence of the human, mouse and rat TH promoters (Kessler et al., 2003; Romano et al., 2005), which revealed that the human TH promoter shares an approximate 46.6% degree of homology with the mouse TH promoter (Romano et al., 2005) and a 30% degree of homology with the rat TH promoter (Gandelman et al., 1990; Kim et al., 2003b). Only five short regions are evolutionarily conserved among the human, mouse and rat TH promoters (Kessler et al., 2003). A human TH minimal promoter encoding for the five CRs placed upstream of the (−194/+35) sequence of the human TH promoter was assembled in a self-inactivating lentiviral vector system and used to transduce GFP into hNPCs and mouse primary striatal and substantia nigra cells, which were in turn treated in vitro with a mixture of differentiating agents to enhance TH gene expression (Romano et al., 2005).
Interestingly, the five CRs could achieve a significant degree of specificity for TH-expressing cells only in hNPCs and not in the mouse cell culture systems (Romano et al., 2005)
Another striking difference in TH gene regulation between the human and mouse TH promoters was observed in a study on NR4A2 (Nurr1), which could not affect in any way human TH gene expression in hNPCs (Jin et al., 2006), in contrast to the mouse and rat systems (Zetterstrom et al., 1997; Castillo et al., 1998; Saucedo-Cardenas et al., 1998; Kim et al., 2002). In addition, a study on the function of three scaffold/matrix attachment regions (S/MARs) present in the promoter at positions (−186/−16) and in the first intron (+645/+755 and +835/+945) of the human TH gene indicated that they might be involved in the differential TH gene regulation among species (Lenartowski and Goc, 2002; Lenartowski et al., 2003). Indeed, different species exhibit a low degree of sequence identity also in the case of the first intron of the TH gene (Kelly et al., 2006). For instance, the first intron of the human and mouse TH genes share only a 50% degree of homology (Kelly et al., 2006). A similar degree of homology was observed for the first intron of human and rat TH genes (Kelly et al., 2006). The role of the three S/MARs in human TH gene regulation was extensively investigated (Meloni et al., 1998; Albanese et al., 2001; Lenartowski and Goc, 2002; Lenartowski et al., 2003). The S/MAR at positions (+645/+755) also encodes for a microsatellite repeat (TCAT) termed HUMTH01, which may act as a regulatory element for TH gene expression (Meloni et al., 1998; Albanese et al., 2001). Interestingly, the microsatellite repeat HUMTH01 is not conserved in the mouse and rat systems, further indicating that the mechanism of TH gene regulation is in fact species-dependent. Regardless of the absence of the aforementioned microsatellite repeat, the first intron of the mouse TH gene seems to be an important mediator for tissue-specific gene expression, as shown by a recent report based on yellow fluorescent protein (YFP) knock-in mice reporter system (Kelly et al., 2006). In this study, YFP was placed in one allele under the control of the endogenous mouse TH promoter via homologous recombination and used to generate a mouse stem (mES) cell line and knock-in mice (Kelly et al., 2006). In these two systems, therefore, YFP replaced the first exon and first intron of the mouse TH gene, while the other allele continued to express endogenous TH. However, YFP and TH gene expression were not completely overlapped both in mES cell line-derived neurons and in knock-in mice brain tissues, in spite of having the entire endogenous mouse TH promoter driving the expression of the reporter gene (Kelly et al., 2006). Per se, such a finding indicates that the deleted region of the mouse TH gene encodes for cis-acting regulatory sequences, which are necessary to confer an accurate tissue-specific TH gene expression (Kelly et al., 2006). All these observations prompted us in engineering the 6.3 kb human TH minimal promoter (−6,244/+35) along with the 1.2 kb first intron fragment of the human TH gene (+730/+1,653) to drive GFP expression in human brain derived cells (Fig. 1). The 1.2 kb first intron fragment contains the microsatellite repeat HUMTH01 and the S/MAR (+835/+945), whereas the 6.3 kb human TH minimal promoter obviously contains the S/MAR (−186/−16). As anticipated, these two genetic elements of the human TH gene optimized GFP expression in human neuroblastoma BE(2)-C-16 cell line (Fig. 2), without, however, conferring specific gene expression only to TH-positive cells (Fig. 3).
Some studies are now focusing on the role of epigenetic programming in the function of TH gene regulation in the human and mouse systems (Aranyi et al., 2002, 2005; Kim et al., 2003c). Intriguingly, a group of investigators found that all dinucleotide CpG sites of the TH promoter are fully methylated in human spermatozoa and in male and female mouse germ cells (Aranyi et al., 2002). In addition to such a paradoxical methylation pattern, the mouse TH promoter exhibited unusual features in preimplantation embryos development, as it remained fully methylated until the morula stage and become demethylated in the majority of cells in the blastocyst (Aranyi et al., 2002).
Further analysis of tissue-specific methylation of the human TH gene revealed the presence of novel regulatory elements in the first exon (Aranyi et al., 2005). The CpG-rich sequence flanking the 5′-end of the human TH gene is very likely to influence the function of the proximal intronic HUMTH01 microsatellite. In this respect, the methylation profile of the CpG island was determined both in TH-expressing and non-expressing cell lines (Aranyi et al., 2005). Indeed, this study revealed a tissue-specific methylation profile, which led to the identification of novel regulatory putative binding sites for AP2, Sp1, and KAISO (Aranyi et al., 2005). The latter is a zinc finger transcriptional suppressor (Daniel and Reynolds, 1999), which binds a consensus sequence containing three methylated cytosines (Prokhortchouk et al., 2001). Three methylated CpG dinucleotides are present in the region (+64/+76) of the human TH gene in cells and tissues that are negative for TH expression (Aranyi et al., 2005). Interestingly, treatment of TH-negative cells with the demethylating agent 5-Azacytidine could increase the fraction of TH-expressing cells in human neuroblastoma CHP212 cell line (Aranyi et al., 2005). Another report showed that histone deacetylase inhibitors such as trichostatin A (TSA) and sodium butyrate induced human TH promoter activity both in neuronal and non-neuronal cell lines (Kim et al., 2003c). All these findings, taken together, indicate that epigenetic programming might play an important role in regulating TH gene expression.
As already mentioned, our data indicated that the region of the first intron (+730/+1,653) displays important functions in optimizing TH gene expression only in brain derived cells, without, however, allowing for specific gene expression in TH-positive cells. Interestingly, Chip analysis was initially conducted with a pan antibody to H4 and found a small cluster of nucleosomes in a region of the promoter, first exon and first intron of the human TH gene both in human neuroblastoma BE(2)-C-16 and human renal carcinoma 293FT cell lines. The latter was used as a cell culture model that is not permissive for TH expression, whereas the former cell line tested positive for TH expression in a 50% cell fraction. Chip analysis with the pan antibody to H4 was carried out to probe the region (−448/+1,204), but the presence of nucleosomes was restricted to the region (−20/+473) in both cell types. This region contains approximately three nucleosomes, as each octameric histone core is wrapped in 1.65 superhelical turns that correspond to 147 bp of genomic DNA (Luger, 2006). However, major differences were observed in terms of H3 and H4 tail acetylation between the two cell lines in the region (−20/+473) of the promoter, first exon and first intron of the human TH gene. All lysine residues of H3 and H4 tails tested positive for acetylation in human neuroblastoma BE(2)-C-16 cell line, whereas the human renal carcinoma 293FT cell line tested positive for acetylation only in the following lysine residues: H3-K14, H3-K23, H4-K5, and H4-K8. Hyper-acetylation of H3 and H4 lysine residues is one of the essential requirements for active transcription, especially if it is observed at regulatory sites upstream of the genes (Litt et al., 2001).
Epigenetic analysis of the promoter, first exon and first intron of the human TH gene might lead to a better understanding of human TH gene regulation in dopaminergic neurons of the substantia nigra. So far, studies that have been conducted point out the importance of the last 3′-end region of the promoter in conjunction with the first exon and first intron in optimizing TH gene expression in brain derived cells and without ectopic expression in other cell types. However, the elements encoded in these genetic regions still do not confer specific gene expression in TH-positive dopaminergic neurons of the substantia nigra. Further studies are indeed necessary to find both other genetic elements of the human TH promoter and factors that mediate the functions of the promoter in regulating TH gene expression. Possibly, these other genetic elements and new factors might work in concert with those already characterized.
The identification of the mechanisms involved in human TH gene expression might lead to the engineering of human TH minimal promoters driving the expression of reporter genes in TH-positive neuronal-based cells, which may derive either from human stem cells of various sources, or hNPCs. Indeed, such a technology might have important implications to implement cell replacement therapeutic approaches for Parkinson's disease.
Acknowledgments
Contract grant sponsor: NIH;
Contract grant numbers: NS24204, NS32519, NS43309.
Literature Cited
- Albanese V, Faucon Biguet N, Kiefer H, Bayard E, Mallet J, Meloni R. Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum Mol Genet. 2001;10:1785–1792. doi: 10.1093/hmg/10.17.1785. [DOI] [PubMed] [Google Scholar]
- Aranyi T, Kerjean A, Toth S, Mallet J, Meloni R, Paldi A. Paradoxical methylation of the tyrosine hydroxylase gene in mouse preimplantation embryos. Genomics. 2002;80:558–563. doi: 10.1006/geno.2002.7011. [DOI] [PubMed] [Google Scholar]
- Aranyi T, Faucheux BA, Khalfallah O, Vodjdani G, Faucon Biguet N, Mallet J, Meloni R. The tissue-specific methylation of the human tyrosine hydroxylase gene reveals new regulatory elements in the first exon. J Neurochem. 2005;94:129–139. doi: 10.1111/j.1471-4159.2005.03173.x. [DOI] [PubMed] [Google Scholar]
- Castillo SO, Baffi JS, Palkovits M, Goldstein DS, Kopin IJ, Witta J, Magnuson MA, Nikodem VM. Dopamine biosynthesis is selectively abolished in substantia nigra/ventral tegmental area but not in hypothalamic neurons in mice with targeted disruption of the Nurr1 gene. Mol Cell Neurosci. 1998;11:36–46. doi: 10.1006/mcne.1998.0673. [DOI] [PubMed] [Google Scholar]
- Daniel JM, Reynolds AB. The catenin p120ctn interacts with Kaiso, a novel BTB/POZ domain zinc finger transcription factor. Mol Cell Biol. 1999;19:3614–3623. doi: 10.1128/mcb.19.5.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Iacovitti L. Protein kinase C activators work in synergy with specific growth factors to initiate tyrosine hydroxylase expression in striatal neurons in culture. J Neurochem. 1997;68:564–569. doi: 10.1046/j.1471-4159.1997.68020564.x. [DOI] [PubMed] [Google Scholar]
- Gandelman KY, Coker GT, Moffat M, O'Malley KL. Species and regional differences in the expression of cell-type specific elements at the human and rat tyrosine hydroxylase gene loci. J Neurochem. 1990;55:2149–2152. doi: 10.1111/j.1471-4159.1990.tb05811.x. [DOI] [PubMed] [Google Scholar]
- Ishiguro H, Arinami T, Saito T, Akazawa S, Enomoto M, Mitushio H, Fujishiro H, Tada K, Akimoto Y, Mifune H, Shiozuka S, Hamaguchi H, Toru M, Shibuya H. Systematic search for variations in the tyrosine hydroxylase gene and their associations with schizophrenia affective disorders, and alcoholism. Am J Med Genet. 1998;81:388–396. [PubMed] [Google Scholar]
- Iwawaki T, Kohno K, Kobayashi K. Identification of a potential Nurr1 response element that activates the tyrosine hydroxylase gene reporter in cultured cells. Biochem Bioph Res Commun. 2000;274:590–595. doi: 10.1006/bbrc.2000.3204. [DOI] [PubMed] [Google Scholar]
- Jin H, Romano G, Marshall C, Donaldson AE, Suon S, Iacovitti L. Tyrosine hydroxylase gene regulation in human neuronal progenitor cells does not depend on Nurr1 as in the murine and rat systems. J Cell Physiol. 2006;207:49–57. doi: 10.1002/jcp.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly BB, Hedlund E, Kim C, Ishiguro H, Isacson O, Chikaraishi DM, Kim KS, Feng G. A tyrosine hydroxylase-yellow fluorescent protein knock-in reporter system labeling dopaminergic neurons reveals potential regulatory role for the first intron of the rodent tyrosine hydroxylase gene. Neuroscience. 2006;142:343–354. doi: 10.1016/j.neuroscience.2006.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler MA, Yang M, Gollomp KL, Jin H, Iacovitti L. The human tyrosine hydroxylase gene promoter. Brian Res Mol Brain Res. 2003;112:8–23. doi: 10.1016/s0169-328x(02)00694-0. [DOI] [PubMed] [Google Scholar]
- Kim JH, Auerbach JM, Rodriguez-Gomez JA, Velasco I, Gavin D, Lumelsky N, Lee SH, Nguyen J, Sanchez-Pernaute R, Bankiewicz K, McKay R. Dopamine neurons derived from embryonic stem cells function in an animal model of Parkinson's disease. Nature. 2002;418:50–56. doi: 10.1038/nature00900. [DOI] [PubMed] [Google Scholar]
- Kim KS, Kim CH, Hwang DY, Seo H, Chung S, Hong SJ, Lim JK, Anderson T, Isacson O. Orphan nuclear receptor Nurr1 directly transactivates the promoter activity of the tyrosine hydroxylase gene in a cell-specific manner. J Neurochem. 2003a;85:622–634. doi: 10.1046/j.1471-4159.2003.01671.x. [DOI] [PubMed] [Google Scholar]
- Kim TE, Park MJ, Choi EJ, Lee HS, Lee SH, Yoon SH, Oh CK, Lee BJ, Kim SU, Lee YS, Lee MA. Cloning and cell type-specific regulation of the human tyrosine hydroxylase gene promoter. Biochem Biophys Res Commun. 2003b;312:1123–1131. doi: 10.1016/j.bbrc.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Kim TE, Park MJ, Hong SJ, Woo MS, Kim SY, Kim KS. Regulation of the tyrosine hydroxylase gene promoter by histone deacetylase inhibitors. Biochem Biophys Res Commun. 2003c;312:950–957. doi: 10.1016/j.bbrc.2003.11.012. [DOI] [PubMed] [Google Scholar]
- Lenartowski R, Goc A. Tissue-specific association of the human tyrosine hydroxylase gene with the nuclear matrix. Neurosci Lett. 2002;330:151–154. doi: 10.1016/s0304-3940(02)00746-2. [DOI] [PubMed] [Google Scholar]
- Lenartowski R, Grzybowski T, Miscicka-Sliwka D, Wojciechowski W, Goc A. The bovine tyrosine hydroxylase gene associates in vitro with the nuclear matrix by its first intron sequence. Acta Biochim Pol. 2003;50:865–873. [PubMed] [Google Scholar]
- Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveals boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Luger K. Dynamic nucleosomes. Chromosome Res. 2006;14:5–16. doi: 10.1007/s10577-005-1026-1. [DOI] [PubMed] [Google Scholar]
- Macaluso M, Cinti C, Russo G, Russo A, Giordano A. pRb2/p130-E2F4/5-HDAC1-SUV39H1-p300 and pRb2/p130-E2F4/5-HDAC1-SUV39H1-DNMT1 multimolecular complexes mediate the transcription of estrogen receptor-alpha in breast cancer. Oncogene. 2003;22:3511–3517. doi: 10.1038/sj.onc.1206578. [DOI] [PubMed] [Google Scholar]
- Macaluso M, Montanari M, Noto PB, Gregorio V, Surmacz E, Giordano A. Nuclear and cytoplasmic interaction of pRB2/p130 and ER-{beta} in MCF-7 breast cancer cells. Ann Oncol. 2006;17:vii27–vii29. doi: 10.1093/annonc/mdl945. [DOI] [PubMed] [Google Scholar]
- Meloni R, Albanese V, Ravassard P, Treilhou F, Mallet J. A tetranucleotide polymorphic microsatellite, located in the first intron of the tyrosine hydroxylase gene, acts as a transcription regulatory element in vitro. Hum Mol Genet. 1998;7:423–428. doi: 10.1093/hmg/7.3.423. [DOI] [PubMed] [Google Scholar]
- Moore RY. Organization of midbrain dopamine systems and the pathophysiology of Parkinson's disease. Parkinson Relat Disord. 2003;9:S65–S71. doi: 10.1016/s1353-8020(03)00063-4. [DOI] [PubMed] [Google Scholar]
- Nagatsu T, Levitt M, Uderfriend S. Tyrosine hydroxylase, the initial step in norepinephrine biosynthesis. J Biol Chem. 1964;239:2910–2917. [PubMed] [Google Scholar]
- O'Malley KL, Anhalt MJ, Martin BM, Kelsoe JR, Winfield SL, Ginns EI. Isolation and characterization of the human tyrosine hydroxylase gene: Identification of 5′ alternative splice sites responsible for multiple mRNAs. Biochemistry. 1987;26:6910–6914. doi: 10.1021/bi00396a007. [DOI] [PubMed] [Google Scholar]
- Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retrovirus by transient transfections. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirih FQ, Tang A, Ozkurt IC, Nervina JM, Tetradis S. Nuclear orphan receptor Nurr1 directly transactivates the osteocalcin gene in osteoblasts. J Biol Chem. 2004;279:53167–53174. doi: 10.1074/jbc.M405677200. [DOI] [PubMed] [Google Scholar]
- Prokhortchouk A, Hendrich B, Jorgensen H, Ruzov A, Wilm M, Georgiev G, Bird A, Prokhortchouk E. The p120 catenin partner KAISO is a DNA methylation-dependent transcriptional repressor. Genes Dev. 2001;15:1613–1618. doi: 10.1101/gad.198501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G, Suon S, Jin H, Donaldson AE, Iacovitti L. Characterization of five evolutionary conserved regions of the human tyrosine hydroxylase (TH) promoter: Implications for the engineering of a human TH minimal promoter assembled in a self-inactivating lentiviral vector system. J Cell Physiol. 2005;204:666–677. doi: 10.1002/jcp.20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. Nurr1 is essential for the induction of the dopaminergic phenotype and survival of ventral mesencephalic late dopaminergic precursor neurons. Proc Natl Acad Sci USA. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soneoka Y, Cannon P, Ramsdale EE, Griffiths JC, Romano G, Kingsman SM, Kingsman AJ. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Wijnen AJ, Bidwell JP, Fey EG, Penman S, Lian JB, Stein JL, Stein G. Nuclear matrix association of multiple sequence-specific DNA binding activities related to SP-1, ATF, CCAAT, C/EBP, OCT-1, and AP-1. Biochemistry. 1993;32:8397–8402. doi: 10.1021/bi00084a003. [DOI] [PubMed] [Google Scholar]
- Yang M, Donaldson AE, Marshall CE, Shen J, Iacovitti L. Studies on the differentiation of dopaminergic traits in human neural progenitor cells in vitro and in vivo. Cell Transplant. 2004;13:535–547. doi: 10.3727/000000004783983729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Dopamine neuron agenesis in Nurr1-deficient mice. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- Zigmond RE, Schwarzschild MA, Rittenhouse AR. Acute regulation of tyrosine hydroxylase by nerve activity and by neurotransmitters via phosphorylation. Annu Rev Neurosci. 1989;12:415–461. doi: 10.1146/annurev.ne.12.030189.002215. [DOI] [PubMed] [Google Scholar]


