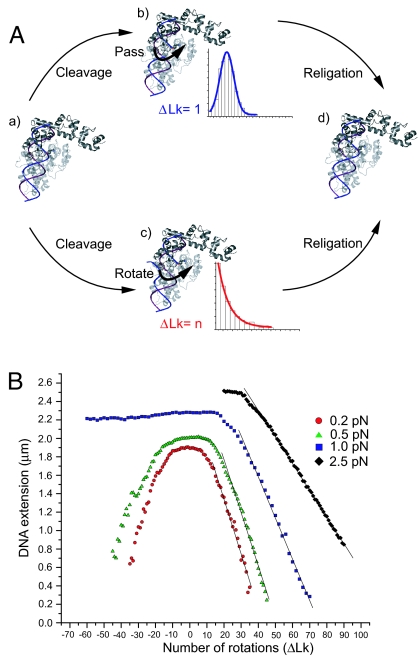
Fig. 1.
Relaxation mechanisms and single DNA molecule calibration. (A) Possible mechanisms of DNA relaxation by topo V. Two possible mechanisms for DNA relaxation have been proposed for type I topoisomerases. Upon DNA binding (a), the relaxation cycle by topo V may proceed either by a strand passage mechanism (b) or a swiveling mechanism (c). The expected step-size distribution is shown for each case. The coordinates of topo-61 (PDB ID code 2CSB) were used to model possible interactions of topo V with DNA (5). (B) DNA calibration. The extension of a 9.7-kb DNA was monitored at different stretching forces. At a low force of 0.2 pN (red circles), the extension of DNA decreases irrespective of the sign of supercoiling because of the formation of plectonemic supercoils. At 0.5 pN (green triangles), DNA extension still decreases irrespective of the direction of rotation of the magnet, although plectonemes coexist with denatured DNA for negative supercoils. At high forces (1 pN, blue squares; 2.5 pN, black diamonds), DNA extension decreases only with the introduction of positive supercoils. The DNA denatures for negative supercoils, and there is no change in DNA extension with introduction of negative rotations. Events only were considered for analysis only when they fell in the linear range of the DNA calibration curve.