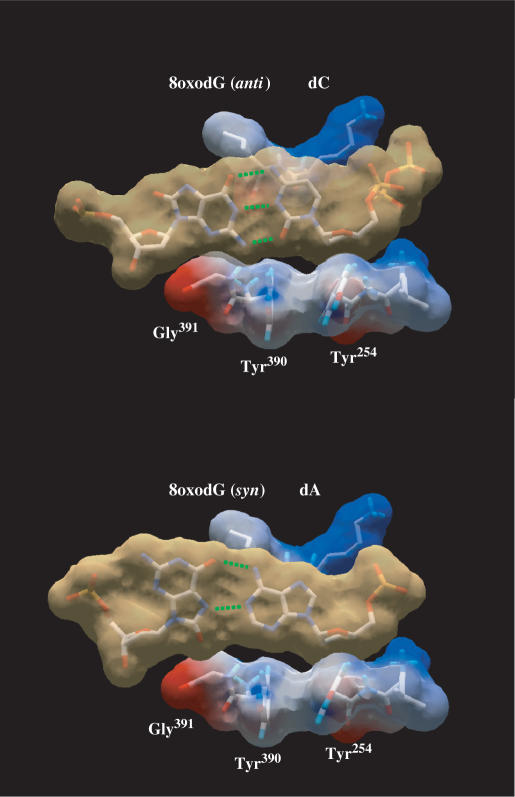
Figure 4.
Modelling of nascent 8oxodG(anti):dC and 8oxodG(syn):dA base pairs in the polymerase active site. Modelling an 8oxodG(syn):dA base pair shows the steric hindrance between the O8 atom and the surface formed by ϕ29 DNA polymerase residues Tyr390 and Gly391, according to (20). van der Waals surfaces are shown for base pairs (light brown) and for protein side chains (coloured according to their electrostaticity). 8oxodG(anti):dC [coordinates obtained from PDB1 Q9Y (20)], and 8oxodG(syn):dA [PDB1 TK8 (19)] base pairs were first modelled into RB69 DNA polymerase active site (PDB1 IG9). Superposition of RB69 and ϕ29 (PDB1 XHX) DNA polymerase active sites allowed us to model both base pairs at the active site of the latter. Green dotted lines represent the potential hydrogen bonds formed between the nucleotides.