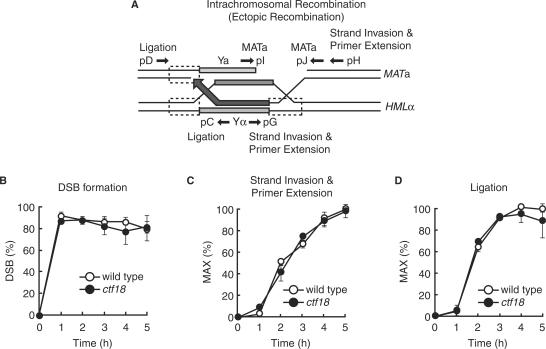
Figure 4.
Ctf18 is not involved in intrachromosomal recombination. JKM161 cells carrying the HML donor sequence were treated as described for Figure 2. (A) A schematic representation of the HR intermediates formed during mating-type switching. The approximate location of the primers used to monitor DSBs (pI and pJ; MATa primers), strand invasion and primer extension (pG and pH; strand invasion and primer extension primers) and completion of repair (pC and pD; ligation primers) are shown. (B, C and D) Genomic DNA was isolated from wild-type (JKM161) and ctf18 mutant (YHO409) cells at the indicated time points after transient expression of HO endonuclease and subjected to PCR analysis to determine the efficiency of DSB formation (B), strand invasion and primer extension (C), and completion of repair (ligation) (C). Amplified PCR products were run on agarose gels and stained with ethidium bromide. DNA signals were quantified and then normalized to the control SMC2 product. A PCR product using primers specific for detecting strand invasion and primer extension will only be generated if invasion of the 3′ strand and replication of at least 222 nt of the donor template sequence occurs. The highest level of strand invasion-primer extension and completion of repair (ligation) in wild-type cells was designated as 100%. The data represent the mean and SD of two independent experiments.