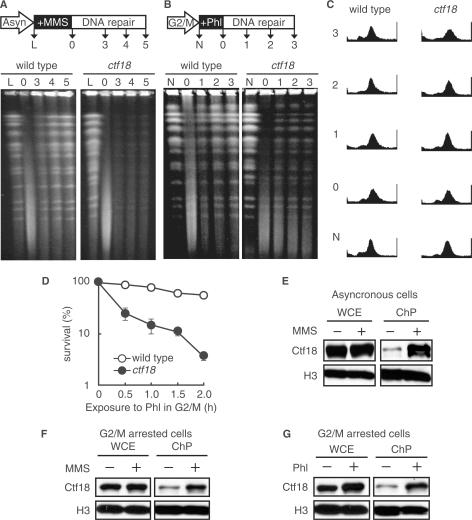
Figure 5.
Association of Ctf18 with the damaged chromatin is required for post-replicative DSB repair. (A) A defect in DNA repair in ctf18 cells was monitored by PFGE. A schematic representation of the process used to prepare the cells in pulsed-field gel electrophoresis (PFGE) is shown (upper panel). Logarithmically growing wild-type (MR101) and ctf18 (YHO218) cells were exposed to 0.1% MMS for 1 h, and then cultured in MMS-free medium for the indicated periods of time. The cells were harvested, and their DNA was analyzed by PFGE as described in the Materials and Methods section. Lane L: untreated cells; Lane 0: cells treated with MMS for 1 h; Lanes 3, 4 and 5: cells treated with MMS for 1 h and then cultured in MMS-free medium for 3, 4 and 5 h, respectively. A representative gel image is shown (bottom panel). (B) Phleomycin-induced post-replicative DSB repair. A schematic representation of the process used to prepare the cells in pulsed-field gel electrophoresis (PFGE) is shown (upper panel). G2/M-arrested cells (wild type; MR101, ctf18; YHO218) were exposed to 100 μg/ml phleomycin at 30°C for 2 h and then cultured in phleomycin-free medium containing 15 μg/ml nocodazole for the indicated periods of time. The cells were harvested and their DNA was analyzed by PFGE as described in the Materials and Methods section. Lane N: untreated cells; Lane 0: cells treated with phleomycin for 2 h; Lanes 1, 2, 3: cells treated with phleomycin for 2 h, then cultured in phleomycin-free medium for 1, 2 and 3 h, respectively. A representative gel image is shown (bottom panel). (C) G2/M-arrested state of (B) confirmed by flow cytometry. To confirm the fact that cells are arrested during the course of above experiment described in (B), we monitored the cell cycle status of cells by FACS. (D) Sensitivity to phleomycin of cells arrested in G2/M. Cells (wild type; MR101, ctf18; YHO218) arrested in the G2/M phase with 15 μg/ml nocodazole were exposed to phleomycin (100 μg/ml) at 30°C for the indicated periods of time, washed to remove phleomycin, inoculated onto YPAD plates containing neither phleomycin nor nocodazole and incubated for 3 days at 30°C before counting the colonies. To measure the sensitivity to phleomycin, at least four cultures were taken for each point. The data represent the means and SD. (E and F) Chromatin binding of Ctf18 after exposure to MMS. Logarithmically growing cells (E) or G2/M-arrested cells (F) expressing Ctf18-13Myc (YHO512) were exposed to 0.1% MMS for 2 h. Aliquots of the cultures were taken before (−) and after (+) exposure to MMS. Whole-cell extracts (WCE) and chromatin containing pellets (ChP) were prepared and analyzed by immunoblotting for the presence of the indicated proteins. Histone H3 was used as a loading control for protein levels. The Myc-tagged proteins and histone H3 were detected using an anti-Myc monoclonal antibody and a specific anti-histone H3 antibody, respectively. (G) Chromatin binding of Ctf18 after exposure to phleomycin. G2/M-arrested cells expressing Ctf18-13Myc (YHO512) were exposed to 100 μg/ml phleomycin for 2 h. Cell aliquots were taken before exposure (−) and 2 h after phleomycin (Phl) exposure (+). Whole-cell extract (WCE) and chromatin containing pellets (ChP) were prepared and analyzed by immunoblot for the presence of the indicated proteins. Histone H3 was used as a loading control for protein levels in WCE and ChP fractions. Ctf18-13myc and Histone H3 were detected using an anti-Myc monoclonal antibody and anti-Histone H3-specific antibody, respectively. To confirm the fact that cells are arrested at G2/M during the course of experiments (F), and (G), we monitored the cell cycle status of cells by FACS (Supplementary Figure S1).