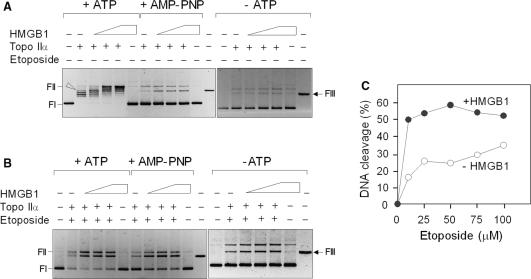
Figure 7.
HMGB1 stimulates DNA cleavage activity of topoisomerase IIα. (A) Effect of HMGB1 on DNA cleavage activity of topo IIα in the absence of etoposide. DNA cleavage reactions contained negatively supercoiled plasmid pBR322 (∼4 nM), human topo IIα (∼8 nM) and increasing concentrations of HMGB1 (1, 2 and 3 μM, left to right). ATP or a non-hydrolyzable ATP analog (AMP-PNP) were at 1 mM. Arrowhead indicates position of linearized DNA. (B) Effect of HMGB1 on DNA cleavage activity of topo IIα in the presence of etoposide. The molar concentrations of topo IIα and plasmid pBR322 were identical as in panel A. HMGB1 protein was at 1 μM and etoposide at 50 μM. (C) Effect of HMGB1 on DNA cleavage by topo IIα in the presence of increasing concentrations of etoposide. Negatively supercoiled plasmid DNA (4 nM) was pre-incubated on ice either with buffer (control) or HMGB1 (1 μM) in the presence of 0–100 μM etoposide. Cleavage reactions were initiated by addition of topo IIα (8 nM) and incubation was carried out at 37°C for 15 min. The cleavage complexes in A–C reactions were trapped by 1% SDS, followed by digestion with proteinase K as detailed in Materials and Methods section. Deproteinized DNA samples were resolved on 1% agarose gels and subsequently stained with ethidium bromide. FIII, HindIII-digested plasmid pBR322 (last right lanes in A and B) indicates mobility of linearized plasmid DNA. FI, supercoiled plasmid DNA; FII, relaxed closed-circular plasmid DNA. The gels are presented as negatives. The percentage of cleaved plasmid DNA was quantified from three independent cleavage experiments, each in duplicates, by ImageQuant TL (GE Healthcare). The average SD for the data was <7%. DNA cleavage is expressed as percentage of initial uncleaved supercoiled plasmid pBR322. (open circle) In the absence (control) or in the presence of HMGB1 (Filled circle).