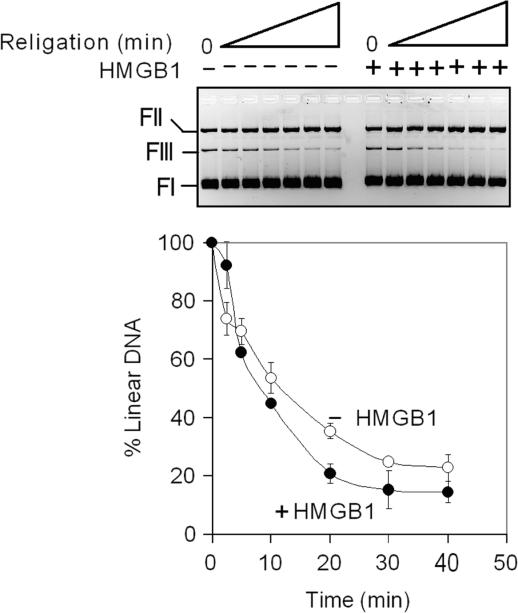
Figure 8.
HMGB1 does not significantly affect DNA religation by topoisomerase IIα. DNA cleavage reactions contained negatively supercoiled plasmid DNA (4 nM), 50 μM etoposide in the absence (control) or presence of HMGB1 (1 μM). Cleavage reactions were initiated by addition of topo IIα (8 nM) and incubation at 37°C for 15 min. The incubation temperature was then shifted to 65°C to suppress cleavage and to promote religation as described in Materials and Methods section. A representative ethidium bromide-stained agarose gel of the DNA religation assay is shown (religation times were 0, 2.5, 5, 10, 20, 30 and 40 min, left to right). The percentage of linear DNA was estimated by the Multi Gauge software using imaging system LAS-3000 (Fuji). Each curve represents the average of 2–3 independent assays. The average SD for the data was <8%. The amount of linearized plasmid molecules in the cleavage reaction containing or lacking HMGB1 was expressed as 100%. (open circle) in the absence (control) or in the presence of HMGB1 (filled circle). FI, supercoiled plasmid DNA; FII, relaxed closed-circular plasmid DNA; FIII, linear plasmid DNA.