Table 1.
Performance and systematic fit of seven models for the eight food web properties
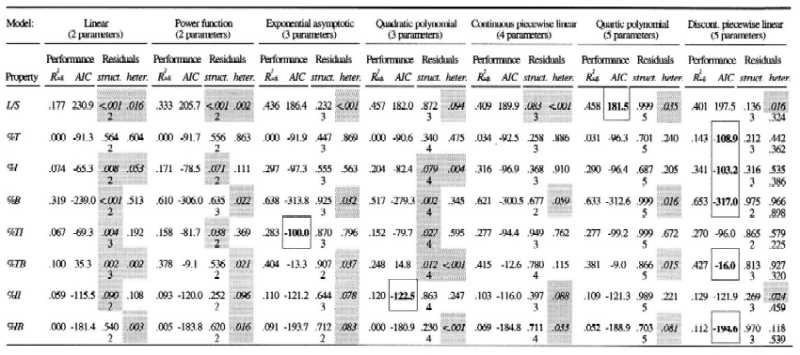
The models investigated are: (i) a linear regression, (ii) a power function, (iii) an exponential asymptotic model, (iv) a second-order polynomial regression, (v) a continuous piecewise linear regression with a breakpoint at c = 12, (vi) a 4th-order polynomial regression, and (vii) a discontinuous piecewise linear regression with a breakpoint at c = 12. The performance of a model is expressed by the adjusted coefficient of determination (Radj2; here, we prefer Radj2 over its unadjusted form, which does not take into account the number of parameters in the model). To determine which model is the most appropriate, given its number of parameters, we computed the Akaike Information Criterion (ref. 27; AIC). The model that minimizes the AIC is considered to be the best one (indicated by boldface type and frame). A systematic error is evaluated in two ways, first by fitting polynomial regressions to the residuals to determine if higher order structure (struct.) is present (see Fig. 1). The P values associated with the F test for the polynomial regressions are given here, together with the order of the polynomial regression applied to the residuals (we tried up to seventh order polynomials; the one giving the lowest P value is shown here). Significant P values (shaded) indicate the presence of systematic pattern in the residuals. Second, we evaluated the presence of heteroscedasticity (heter.; see Fig. 3) by fitting a linear regression to the absolute values of the residuals (28). The P values associated with the F test are given here. A poor fit to the linear regression indicates the absence of heteroscedasticity. For the piecewise model, the first P value is for webs with 12 species or less, and the second is for larger webs. As an example, the discontinuous piecewise linear model explains 40.1% of the variance in the link density property L/S, and gives an AIC of 197.5; a cubic regression of the residuals on S gives a P value for the F test of 0.136; a regression of the absolute values of the residuals on S gives a P value of 0.016 for the webs with 12 species or less and a P value of 0.324 for the webs with >12 species.
