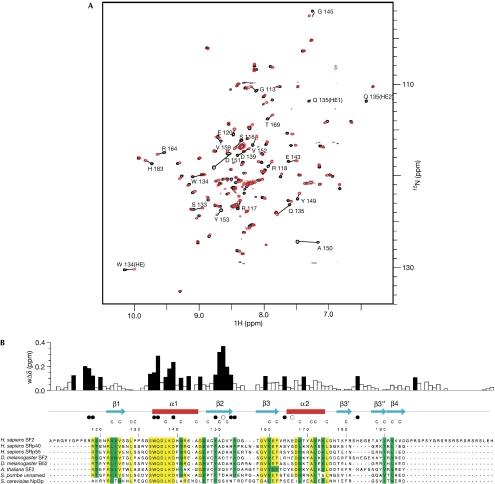
Figure 2.
Interaction of SF2/ASF RNA-recognition motif 2 with RNA. (A) Chemical shift perturbation following the addition of 5′-ACGA RNA to 15N-labelled RRM2. The heteronuclear single quantum correlation (HSQC) spectrum of free protein (black) and after the addition of 1.75 molar equivalents of RNA (red) are shown. Amino acids showing marked changes are indicated. (B) Magnitude of weighted amide chemical shift perturbations (wΔδ) following the addition of 5′-ACGA RNA to 15N-labelled RRM2. Mapping of these perturbations to secondary structural elements are shown above the SF2/ASF sequence alignment with related proteins. Residues with chemical shift changes of more than 0.1 p.p.m. are shown by black bars. The side chain Hɛ resonance of W134 also shows a sizeable shift of more than 0.1 p.p.m., and the side chain Hɛ of Q135 is unobservable, following the first addition of ligand, which indicates exchange broadening owing to a large chemical shift change. Filled circles below the secondary structural elements represent amino acids directly involved in RNA binding identified by mutagenesis, and open circles represent amino acids in which mutagenesis does not affect RNA binding. Residues that form the core of the RRM fold are denoted by the letter C above the sequences. Within the sequence alignments, residues highlighted in yellow are strongly conserved and residues in green have conservative substitutions. RRM, RNA recognition motif; SF2/ASF, splicing factor 2/alternative splicing factor.