Figure 7. Analysis of RACK1, c-Jun, cyclin D, P-ERK and P-JNK levels in melanoma tumor samples.
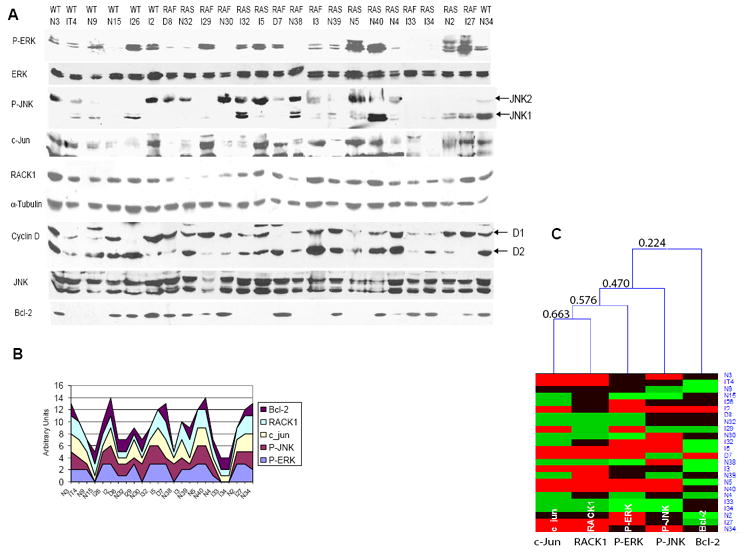
A.ERK activity is associated with JNK, c-Jun and RACK1 levels and activity in human tumor samples. Protein samples (80μg) obtained from melanoma tumors were analyzed by Western blot using the indicated antibodies. For the RACK1, Bcl-2 and α-Tubulin blots, 20μg of protein was used. The status of B-RAF and N-RAS genes is shown above the tumor ID. RAS indicates a Q61K or Q61R mutation. RAF indicates a V599E mutation. WT indicates the absence of Q61 and V599 mutation in N-RAS or B-RAF.
B. Stack graph represents relative levels of P-ERK, c-Jun, RACK1 and P-JNK that were quantified (Fig. S16) and converted into numbers (0, 1, 2, 3) to reflect protein expression level qualitatively.
C. Hierarchical clustering of the samples shown in A was performed with the aid of HCE program (Seo et al., 2002), using average linkage and Euclidean distance measure between expression levels. Different expression levels are shown in different colors: 0- light green, 1- green, 2- black and 3- red. The order of linking corresponds to the proximity of expression levels in different cell types. Minimal similarity values (computed in HCE program using Euclidean distance and normalized to a scale from 0 (no similarity) to 1 (identical profile)) are shown on the tree to reflect qualitatively similarities in expression patterns. The analysis revealed significant relations between P-ERK and c-Jun (0.672), c-Jun and RACK1 (0.617) and between P-ERK and RACK1 (0.680) levels. These values are significant with 99% confidence (the critical value for 1% significance level for 22 degrees of freedom (24-2) is 0.515, using Student’s t-statistics). Bcl2 levels, which are not directly associated with the other proteins in this analysis, served as an outlier for the statistical analysis.
