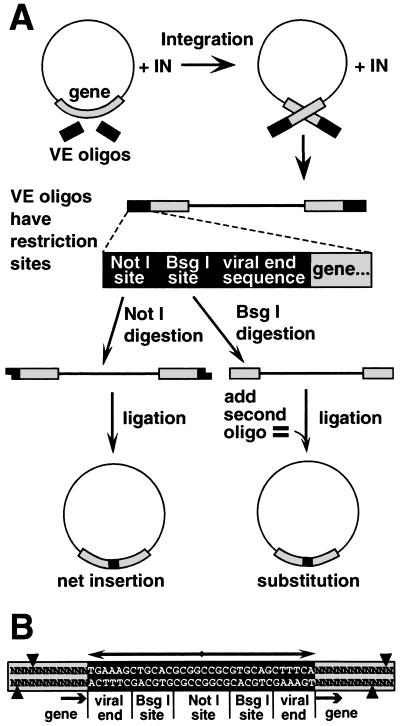
Figure 2.
Strategy for making random insertions. (A) MoMLV integrase inserted oligonucleotide duplexes (VE oligonucleotides) containing terminal sequences from MoMLV DNA ends into diverse sites in a plasmid. Linear DNA products of concerted integration of the VE oligonucleotides were purified and digested with NotI. Intramolecular ligation of the resulting cohesive ends produced a 36-bp insertion in the gene, or elsewhere in the plasmid. Digestion of the concerted integration product with BsgI (lower right) resulted in excision of the integrated oligonucleotide duplex, along with 12 bp of the flanking DNA. (B) Sequence of the insertion mutation and flanking target DNA. Each insertion has a stereotyped structure composed of a 32-bp palindrome (double-headed arrow), containing a NotI site, two BsgI sites, and two copies of sequences representing the ends of MoMLV DNA. Arrows mark the integrase-generated four-base duplication of target DNA sequence. Cleavage directed by the two BsgI sites is depicted by arrowheads, and typically resulted in a 12-bp deletion in the target gene.