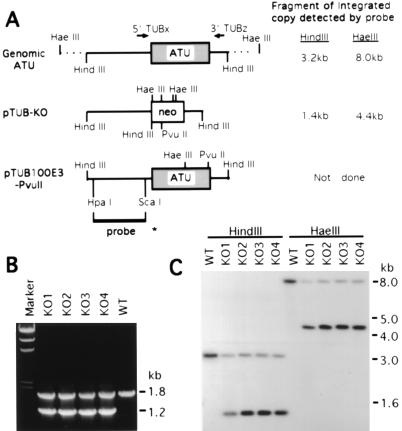
Figure 2.
(A) Maps of the macronuclear ATU gene and the transforming constructs, pTUB-KO and pTUB100E3-PvuII. The 1.4-kb ATU coding region is shown as a shaded box in an 8.0-kb HaeIII genomic fragment. In the knockout construct pTUB-KO, the 0.8-kb neo coding region (shown as an open box) contains four HaeIII sites, a HindIII site, and a PvuII site. pTUB100E3-PvuII contains a 3.2-kb cloned HindIII fragment of the ATU gene which is silently marked by two restriction sites, HaeIII and PvuII. The HpaI/ScaI fragment used as an ATU probe in the Southern blot analysis is shown as a bold line with an asterisk at its end. Fragment sizes of integrated genomic copies produced by digestion with HindIII or HaeIII which hybridize to the 5′ ATU probe are indicated, as are the sites of the PCR primers used in B. (B) PCR analysis of the ATU gene in four pTUB-KO somatic transformants (KO1–4). 5′TUBx and 3′TUBz (locations shown in A) were used as primers in PCR. The 1.8-kb band detected in wild type (WT) and in KO1–4 transformants is derived from the endogenous wild-type ATU gene. The 1.2-kb band is the size expected for the replacement of the wild-type gene by the knockout construct containing the neo gene. Marker, HindIII-cut λ DNA. (C) Southern blot analysis of the pTUB-KO transformants. Digestion with HaeIII is used to assay for homologous recombination, while HindIII digestions served as controls. When genomic DNA is digested with HaeIII, the 8.0-kb fragment derived from the wild-type ATU locus is detected in wild-type control (WT) as well as in KO1–4; the 4.4-kb band corresponds to the same locus when the neo gene replaces the ATU coding region. When digested with HindIII, the 3.2-kb band derived from the wild-type ATU gene is detected in all clones; the 1.4-kb band corresponds to the replacement fragment containing the neo gene.