Fig. 1.
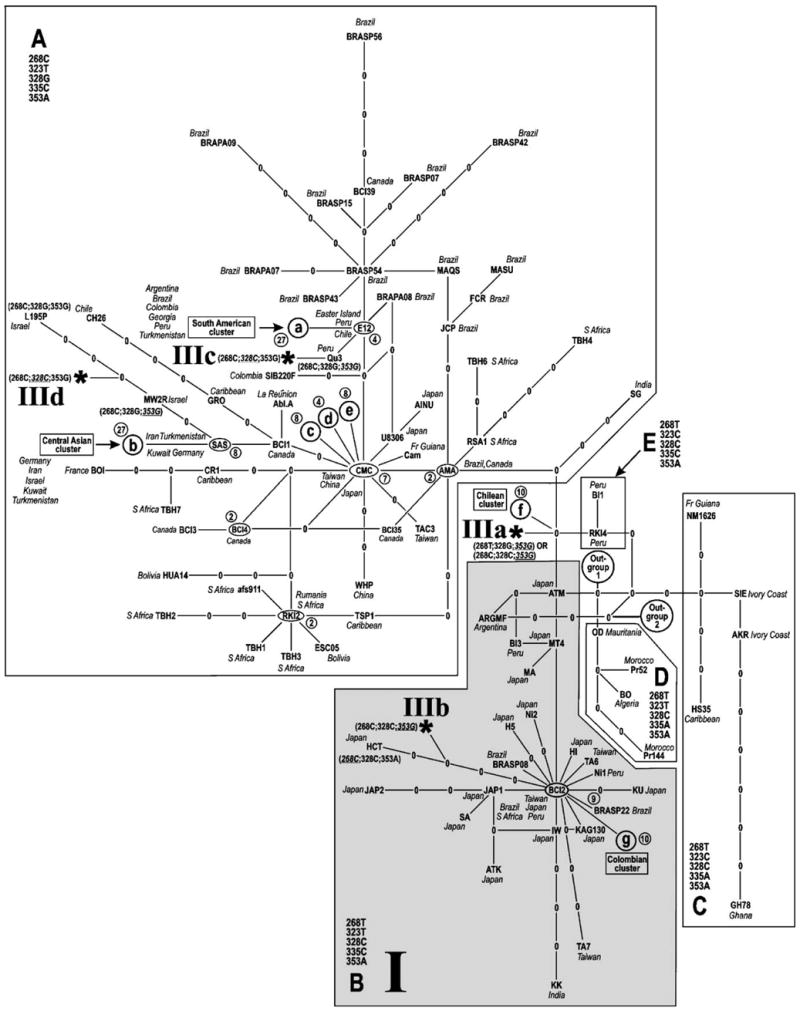
Cladogram of 168 strains belonging to the Cosmopolitan clade of HTLV-1, estimated using statistical parsimony (Templeton et al., 1992) as implemented in the program TCS, version 1.13 (Clement et al., 2000) and a multiple-sequence alignment of 521 bp from the LTR region. Branch tips and internal nodes occupied by extant viral sequences are labelled by the strain name, empty internal nodes by zeroes. Nodes co-occupied by more than one strain with the same genotype are indicated by ellipses, labelled with a single representative strain name from the co-occupying set. Numbers of co-occupant strains at such a node are shown in circles adjacent to each such node. Each line segment, regardless of length as drawn, represents one inferred mutational step as defined by a 95% confidence limit of nine steps. Closed cells in the network result from multiple, equally parsimonious connections between subnetworks at a particular step during execution of the TCS algorithm. For visual clarity locations of five strain clusters are symbolized schematically: (a) South American cluster; (b) Central Asian cluster; (c–e) miscellaneous strains. Two equally parsimonious attachment points of an outgroup composed of 24 non-Cosmopolitan (African and Melanesian) strains to the main network are labelled. Subclades A–E are demarcated in boxes. Predominant nucleotide states found within each subclade at five phylogenetically informative positions (268, 323, 328, 335 and 353) are indicated. Where the genotype of an individual strain differs from the predominant state within a subclade, only the states of divergent sites are given. Occurrence of mummy-associated LTR genotype I (Li et al., 1999) throughout subclade B is indicated by gray shading and the Roman numeral “I”. Possible phylogenetic placements of mummy-associated LTR sequences belonging to genotype III (Li et al., 1999) are marked by asterisks and labelled IIIa–IIId. Note that points of attachment IIIb and IIId of mummy genotype III on the unoccupied branches leading to strains HCT, MW2R and L195 cannot be specified exactly; one possibility was therefore chosen arbitrarily in each case. Nucleotide positions are numbered according to reference strain ATK (GenBank J02029).
