Abstract
The signal-recognition particle (SRP) is a ubiquitous protein–RNA complex that targets proteins to cellular membranes for insertion or secretion. A key player in SRP-mediated protein targeting is the evolutionarily conserved core consisting of the SRP RNA and the multidomain protein SRP54. Communication between the SRP54 domains is critical for SRP function, where signal sequence binding at the M domain directs receptor binding at the GTPase domain (NG domain). These SRP activities are linked to domain rearrangements, for which the role of SRP RNA is not clear. In free SRP, a direct interaction of the GTPase domain with SRP RNA has been proposed but has never been structurally verified. In this study, we present the crystal structure at 2.5-Å resolution of the SRP54–SRP19–SRP RNA complex of Methanococcus jannaschii SRP. The structure reveals an RNA-bound conformation of the SRP54 GTPase domain, in which the domain is spatially well separated from the signal peptide binding site. The association of both the N and G domains with SRP RNA in free SRP provides further structural evidence for the pivotal role of SRP RNA in the regulation of the SRP54 activity.
The signal-recognition particle (SRP) targets proteins destined for membrane insertion and secretion to or across the plasma membrane in bacteria and the endoplasmic reticulum in eukaryotes. SRP binds to the ribosome as well as to the hydrophobic signal sequences of nascent polypeptide chains as they emerge from the ribosome. The ribosome-nascent chain–SRP complex is then targeted to the membrane by an interaction between SRP and its receptor (SR in eukaryotes and FtsY in bacteria). In the presence of the translocon, the signal peptide is released into the translocon channel. SRP then dissociates from SR and the ribosome-nascent chain and is ready for another cycle of cotranslational protein targeting. The targeting cycle is coordinated by GTP binding and hydrolysis by both SRP and SR (for reviews, see refs. 1–3).
SRP composition varies across the three domains of life. Mammalian SRP consists of a single ≈300-nt RNA (SRP RNA or 7S RNA) and six proteins, which can be divided into two major functional domains: the Alu domain (comprising the proteins SRP9 and -14) and the S domain (SRP19, -54, -68, and -72). The S domain functions in signal sequence recognition and SR interaction, whereas the Alu domain is required for translational arrest on signal sequence recognition (4). Archaeal SRPs consist of a 7S RNA that is highly similar to mammalian 7S RNAs, but only two homologues of the mammalian SRP proteins, namely SRP19 and SRP54 (5). A minimal set of SRP components is found in bacteria, which consist of shorter RNAs (4.5S RNA) and only Ffh, the protein homologous to SRP54 (6, 7).
SRP54, the only protein component present in all SRPs, comprises an N-terminal domain (N, a four-helix bundle), a central GTPase domain [G, a ras-like GTPase fold, with an additional unique α-β-α insertion box domain (IBD)], and a methionine-rich C-terminal domain (M, an all-α structure) (8, 9). The N and G domains are structurally and functionally coupled; together, they build the NG domain that is connected to the M domain through a flexible linker (10). The M domain anchors SRP54 to SRP RNA and carries out the principal function of signal sequence recognition (11–13). The NG domain interacts with the SR in a GTP-dependent manner (14, 15).
Biochemical studies (16–19) and the x-ray structures of free SRP54 (10, 20), SRP54 in complex with helix 8 (20), the Ffh NG domain/FtsY NG domain complex (14, 15), and the cryo-EM structures of ribosome-bound SRP (21–23) show major structural flexibility within SRP54. It is suggested that the conformational changes in SRP54 are fundamental to the coordinated binding and release of the ribosome, the signal peptide, the SRP receptor, and the translocon. Further evidence for this comes from the structures of the free Sulfolobus solfataricus SRP54–helix 8 complex (20) and the mammalian SRP bound to signal peptide and ribosome (21–23). These show different relative orientations of the NG domain at two different stages in the SRP cycle. In both the mammalian and S. solfataricus structures, SRP54 assumes a so-called “open” conformation, in which the NG domain and the RNA are separated by at least 20 Å. Biochemical data, however, indicate that, in free SRP, the NG domain can directly interact with SRP RNA (17, 18, 24–26). To further address this discrepancy and to better delineate the structural basis of the first step in the SRP cycle, we have determined the crystal structure of the free S domain of SRP of the Archaeon Methanococcus jannaschii at 2.5-Å resolution. The structure presented in this paper reveals a direct interaction between the SRP54 NG domain and SRP RNA and highlights aspects of the initiation of the signal peptide recognition event and its control.
Results
Identification of an RNA-Bound SRP54 NG Domain Conformation.
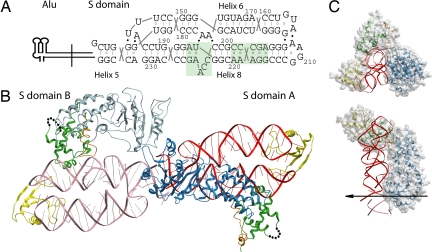
The crystals of the S domain complex of M. jannaschii SRP, consisting of the S domain RNA (7S.S RNA, nucleotides G142–G237), SRP19, and SRP54, diffracted to 2.5-Å resolution. The structure was determined by molecular replacement (Table 1). The asymmetric unit comprises two S domain complexes, termed A and B, that are joined by end-to-end packing of RNA helix 5 such that a pseudocontinuous helical element is formed (Fig. 1). Helices 6 and 8 lie side by side, with helix 8 coaxially stacked onto helix 5, thereby forming a continuous helical subdomain that connects to helix 6 via the three-way junction. The α/β structure of SRP19 binds to the tetraloop regions of helices 6 and 8 and forms a hydrogen-bonding network identical to the one described for the 7S.S RNA–SRP19 complex structure in ref. 27. The helix–turn–helix motif in the SRP54 M domain (Gly-324-Leu-427) binds to the widened minor groove of mismatches in the symmetric loop and the invariant A195 in the asymmetric loop of helix 8. The intermolecular contacts between the M. jannaschii M domain and these RNA loops are remarkably conserved to those in the Escherichia coli M domain–SRP RNA (28) and the human M domain–SRP19–SRP RNA (29) complexes. Notably, in the M. jannaschii complex, the M domain and SRP19 are in direct contact by hydrogen-bonding and stacking interactions. These are located between Arg-401 in the M domain and residues His-57–Trp-58 situated in the helix-8 binding loop L3 of SRP19 and explain the detrimental effect of a His57Ala/Trp58Ala double mutant on SRP54 assembly (30).
Table 1.
Data collection and refinement statistics
| Data collection | |
| Wavelength, Å | 0.979 |
| Resolution range, Å | 30.0–2.5 (2.6–2.5) |
| Total number of observations | 277,638 |
| Unique reflections | 49,835 (7,094) |
| Completeness, % | 99.1 (95.2) |
| Redundancy | 5.6 (5.0) |
| 〈I/σ(I)〉 | 6.3 (1.6) |
| Rmerge, % | 12.6 (76.5*) |
| Refinement | |
| Resolution range, Å | 20.0–2.5 |
| Reflections in Rwork | 46,767 |
| Reflections in Rfree | 2,620 |
| Rwork, % | 24.4 |
| Rfree, % | 29.4 |
| rms deviations | |
| Bond lengths, Å | 0.021 |
| Bond angles, ° | 2.953 |
| Torsion angles, ° | 21.895 |
| No. of atoms | 12,353 |
| RNA | 4,126 |
| Protein | 7,736 |
| Water molecules | 474 |
| Mean temperature factor for all atoms, Å2 | 50.4 |
| SRP19, S domain A, B | 39.2, 39.8 |
| SRP54, S domain A, B | 54.6, 63.2 |
| SRP RNA, S domain A, B | 32.1, 35.3 |
| Water molecules | 45.9 |
Values in parentheses are for the highest-resolution shell.
*The R factor for the highest-resolution shell is high; however, the redundancy of 5.0 allows useful data to be obtained in this shell.
Fig. 1.
Overall structure of the SRP54–SRP19–7S.S RNA complex. (A) The nucleotide sequence and two-dimensional topology diagram of the 7S.S RNA fragment used for crystallization are given with the numbering corresponding to full-length M. jannaschii 7S RNA. The lines between bases indicate Watson–Crick base pairs, the open circles indicate noncanonical pairs, and the filled circles indicate tertiary RNA–RNA interactions. Highlighted in green are the symmetric and asymmetric loops in helix 8. (B) Ribbon representation of the asymmetric unit of the crystal, which contains two S domains. Their respective RNAs are stacked end to end, forming a pseudocontinuous helical element. In S domains A and B, the NG domain is shown in blue and light blue, and 7S RNA is shown in red and light red, respectively; the M domain is shown in green; the GM-linker is shown in orange; and SRP19 is shown in yellow. The 25-residue finger loop (indicated by dots) in the M domains was not observed in the electron density maps. (C) Top and side view of the structure of S domain A. The molecular surface of the NG domain highlights the good fit with 7S.S RNA. The noncrystallographic twofold axis is indicated by an arrow. The images are color-coded as in A.
Whereas the orientation of the 7S.S RNA, SRP19, and the M domain is almost identical in the two S domains of the asymmetric unit, the orientation of their respective NG domains (Met-1–Asp-298) differs significantly. In S domain A, the NG domain and the RNA are closely associated, and direct protein–RNA contacts are formed between both the N and G domains with the RNA: the N domain interacts with the distal helix 8 and the G domain interacts with the proximal helices 8 and 6 and the adjoining helix 5. The G domain's helix 5 interface in S domain A extends beyond its own helix 5, into the helix 5 in S domain B (helix 5′). As a consequence, the NG domain of B cannot occupy the same position as in S domain A because of steric clashes between the two G domains. In B, the NG domain is tilted and rotated around helix 8 into an open conformation showing no contact between it and the RNA (Fig. 1). This NG domain movement is linked to conformational changes localized to residues Leu-293–Gly-324 that connect the G and M domains. The NG domain of A is well ordered in the crystal. By comparison, the electron density for the NG domain of B is weaker, and its position is consequently less well defined.
Induced Conformational Changes in 7S.S RNA During SRP54 Assembly.
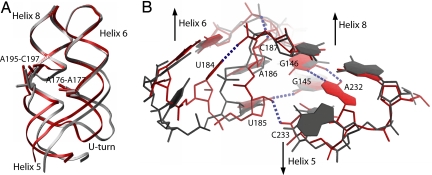
It has been shown previously that the asymmetric loop in helix 8 is flexible and undergoes critical structural changes on binding of the SRP54 M domain (20, 28, 29). In free M. jannaschii 7S.S RNA, the three unpaired bases 195-ACC in the asymmetric loop are pointing toward the interior of the RNA helix (30). Binding of SRP19 to the tips of helices 6 and 8 causes a backbone inversion in the 195-ACC bulge so that its bases become splayed out on the surface of the RNA helix (27). The structure of the ternary complex presented here reveals that, on interaction with SRP54, these bases become stably stacked forming a structure similar to the “RNA platform” observed in bacterial and human SRP (28, 29). Importantly, the formation of the RNA platform involves an upward movement of the bulged backbone. This movement brings the asymmetric and symmetric loops into close proximity, forming the conserved contact between 2′-OH of A195 and the phosphate oxygen of A219. Thus, SRP54 M domain binding causes a shortening of helix 8 and a reorientation of its proximal part, consequently displacing the nucleotides at the SRP54 G domain binding site by up to 7 Å (Fig. 2A). A similar structural change has been observed in human SRP RNA upon M domain binding (29). The asymmetric loop and helix 6 are in direct contact via A-minor-type interactions formed between the G-C and reversed A-U base pairs flanking the 195-ACC bulge and the looped-out adenosines, A176 and A177. These tertiary contacts play an important role in communicating conformational changes from the SRP19 binding site to the asymmetric loop (27, 30). Similarly, they convey the M domain-induced conformational changes from the asymmetric loop to helix 6. With the distal parts of helices 6 and 8 now held in place by SRP19, the altered positions of A176 and A177 enforce changes in the proximal part of helix 6 containing the U-turn loop. Newly formed RNA–RNA interactions near the three-way junction are shown in Fig. 2B. Elevated temperature factors of the nucleotides in helices 6 and 8 and helix 5 in the SRP19–RNA complex indicate intrinsic flexibility likely to be required for SRP54 recognition (30). As expected, in the ternary complex, these nucleotides are highly ordered with low-temperature factures and well defined electron densities [supporting information (SI) Fig. 6].
Fig. 2.
Induced conformational changes in 7S.S RNA during SRP54 assembly. Shown is the superposition of 7S.S RNA in complex with SRP54–SRP19 (S domain A, red) and with SRP19 only (gray). (A) Conformational changes in the proximal part of 7S.S RNA are initiated at the asymmetric loop of helix 8 and conveyed to helix 6 via adenosines A176 and A177. The concerted movement of helices 6 and 8 makes nucleotides in helix 5 shift by up to 7 Å. The same movement is seen in S domain B, showing that this structural change in the RNA is caused by the RNA–M domain interaction. The superposition is based on SRP19; SRP19 and SRP54 are not shown for clarity. (B) RNA–RNA stabilization in the proximal part of 7S.S RNA. Newly formed hydrogen bonds (dotted lines) in the ternary complex include a hydrogen bond from the N3 group of U184 to the phosphate moiety of C187 internally stabilizing the U-turn. U185 becomes part of a base triple with the G145–C233 base pair situated immediately below the three-way junction. Furthermore, the riboses of U185 and A186 are turned toward the minor groove of helix 5 and form tertiary interactions with the 2′-OH atoms of C233 and G146. Nucleotide A232, which is looped out in the absence of SRP54, becomes stably stacked into the RNA helix to form a heteropurine G–A base pair with G146 in the junction. The superposition is based on phosphate atoms of nucleotides G230–C236.
RNA Binding Involves Residues from Both the N and G Domains of SRP54.
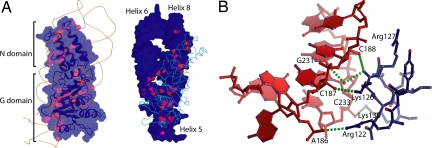
The NG domain in S domain A associates lengthwise with the 7S RNA, burying 2,200 Å2 of solvent-accessible surface area (Fig. 3A). This makes the NG domain–RNA interface considerably larger than the 1,350-Å2 M domain–RNA interface. However, M domain recognition is mediated by the formation of an intricate intermolecular interface, including seven base-specific interactions. In contrast, the NG domain makes relatively few direct contacts and those that are formed are exclusively with the sugar–phosphate backbone. The RNA-binding surface of the NG domain does not overlap with the SR-binding surface (14, 15) and involves predominantly the loops in the G domain that are unique to the SRP-type GTPases (SI Fig. 7). These loops pack with a good shape complementarity against the proximal parts of helices 6 and 8, and helices 5 and 5′. The G domain loops in contact with helix 5′ include the loop forming the extended linker connecting the N and G domains (Leu-86–Gln-100) and the loop connecting α-helix αG3 in the IBD domain to β-strand βG4 (Phe-179–Asp-183). Here, the side chain of Lys-99 forms a hydrogen bond to the phosphate oxygen of C233′, and the tandem lysine residues Lys-180 and Lys-181 interact with the phosphate oxygen of A232′ and the 2′-OH group of C188′ situated across the minor groove. The loop connecting the secondary structural elements, αG1 and βG2, is tightly inserted into the helix 5 minor groove (Fig. 3B). The C-terminal helix of the G domain is situated in the NG–RNA interface and directs the side chains of Lys-288 and Arg-292 toward helices 8 and 6 (SI Fig. 8). Positively charged residues in the αG1–βG2 loop and the C-terminal helix are conserved in SRP54, but not in the non-RNA-binding SR homologues (5).
Fig. 3.
SRP54 NG domain–RNA interactions. (A) The molecular surface of the SRP54 NG domain (Left) and 7S.S RNA (Right) are shaded to indicate the different accessibilities of the surface areas at each residue (Left) and nucleotide (Right) between the free and complexed forms. The red areas define protein–RNA contacts. The molecule to the right is rotated by 180° with respect to the molecule to the left. (B) Interaction between the G domain loop connecting αG1 and βG2 and the RNA minor groove of helix 5. The side chains of Arg-122 and Lys-126 bind to the phosphate oxygen of A186 and the 2′-OH atom of C187 in the RNA strand that switches from helix 6 to 8. The main-chain oxygen of Lys-126 is hydrogen-bonded to the 2′-OH of C188, and forms a water-mediated contact with the guanine base of G231, the base partner of C187 in the G–C pair immediately above the three-way junction. Furthermore, the side chain of Lys-130 interacts with the phosphate group of C233. RNA is colored in red, protein residues are colored in blue, and hydrogen bonds are colored in green.
Direct contacts between the four-helix bundle N domain and 7S RNA are made at the tip of the N domain. Here, side chains of residues in αN1 and αN4, situated next to the two apical loops (Ala-18–Lys-24 and Lys-63–Ser-69), are in close proximity to the RNA backbone of the distal helix 8. Arg-17 forms a hydrogen bond to the 2′-OH group of G213 in the tetraloop-closing G-C pair, and Lys-71 and Glu-72 form hydrogen bonds with the backbone of nucleotides G204 and A203 in the symmetric loop.
The Linker Between the NG and M Domains Is Dynamic.
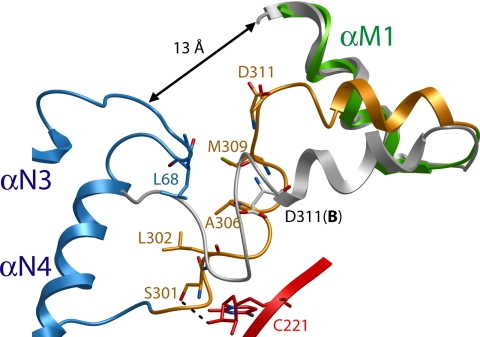
The SRP54 structures in S domain A and B start to deviate from each other at residue Leu-293 situated in the conserved 292-RLLGMGD sequence motif at the C terminus of the G domain. This motif has previously been identified to form a hinge region in SRP54 (20). In S domain A, the motif forms a sharp αβ-turn, which is packed between the N domain and helix 8 (SI Fig. 9). The succeeding 25 residues constituting the GM-linker (Leu-299–Arg-323) are not well defined in the electron density, and their elevated temperature factors suggest linker flexibility in both S domains A and B. In A, the linker assumes an overall “L-shaped” conformation. The N-terminal arm (Leu-299–Val-310) adopts a distorted helical structure. Residues within this linker segment are in contact with helix 8 and the apical loop of the N domain (Fig. 4). After a 90° bend at residue Asp-311, the C-terminal arm (Glu-312–Arg-323) forms a short α-helix, which packs against αM1 lining the hydrophobic groove in the M domain. In B, the contacts made by the linker residues with the RNA and the N domain loop are lost, which results in unbending at Asp-311 and an extension of the α-helix by one turn. The SRP54 M domain structures in A and B superimpose well from the conserved 324-GKFTL sequence motif, which is in close contact with the M domain's C-terminal helix and hydrophobic core. The α-helix preceding the 324-GKFTL loop is oriented differently in A and B and thus packs against αM1 at different angles with distinct packing interactions. This observation indicates a hinge region at the 324-GKFTL loop and supports the notion that the GM-linker–αM1 interface can be smoothly adjusted (20). Importantly, in S domains A and B, no contact exists between the NG and M domains apart from their covalent linkage in the linker sequence. In A, the N and G domains have a center-of-mass distance from the M domain of 36 and 55 Å, respectively. A minimum gap of 13 Å separates the main chains of the apical loops in the N domain from the hydrophobic groove in the M domain.
Fig. 4.
Ribbon representation of the GM-linker structure. In S domain A, Ser-301 forms a hydrogen bond with the 2′-OH group of C221 in helix 8, and Leu-302, Ala-306, and Met-309 make hydrophobic interactions with Gly-67 and Leu-68 situated in the apical loop between αN3 and αN4 in the N domain. The NG domain is shown in blue, the RNA is shown in red, the M domain is shown in green, and the GM-linker is shown in orange. The GM-linker in S domain B is shown in gray. The side chain of Asp-311 in B is also shown. The overlay is based on the SRP54 M domains.
Discussion
The structure of the SRP54–SRP19–7S.S RNA complex from M. jannaschii has been characterized in its free state. In the crystal, the asymmetric unit contains two S domain complexes with different SRP54 NG domain orientations. One NG domain is bound to 7S RNA forming an overall compact and ordered S domain structure (S domain A). The other NG domain is separated from the RNA forming an open structure, in which the position of the NG domain is not well defined (S domain B). Because the NG domain RNA-binding site also includes nucleotides from the neighboring RNA in the asymmetric unit, steric clashes prevent the two NG domains from binding the RNA at the same time and may explain the flexibility of the NG domain in S domain B. The previously described S. solfataricus SRP54–helix 8 structure reports an open complex structure (20). However, the NG domain orientation in this structure differs significantly from the open structure observed in S domain B (SI Fig. 10).
A direct interaction between the NG domain and the SRP RNA in free SRP has been suggested by prior biochemical studies (17, 18, 24–26). The NG–RNA interactions observed in S domain A are consistent with some but not all of these data. RNA protection and cross-linking studies identified regions in E. coli 4.5S RNA proximal to the domain IV, the homologue region to helix 8, as the binding site for the NG domain (17, 24). Consistent with the proposed binding site, the isolated NG domain shows much weaker binding to a 49-mer RNA, which comprises only domain IV, than to full-length 4.5S RNA (17). The NG domain binding site found in S domain A agrees with the binding site proposed by these biochemical findings. The absence of most of the G domain binding site in the 45-mer RNA used for crystallization of the S. solfataricus SRP54–helix 8 complex thus may explain the observed open non-RNA-bound structure in this case. However, a high dissociation constant of the NG domain for 4.5S RNA (17) and weak RNA footprints produced by the NG domain, together indicate that the NG–RNA interaction is weak and might be more dynamic in free SRP. Hence, it is possible that the different NG domain orientations observed in S. solfataricus and M. jannaschii SRP represent examples from a dynamic range or discrete states of conformations that occur in free SRP. Structure-guided experiments with free SRP are currently being pursued to address this possibility. A number of results, however, support the biological relevance of the SRP54-SRP RNA interactions observed in S domain A, as will be further discussed.
In the first step of cotranslational protein targeting, the SRP binds to the signal sequence of the nascent polypeptide chain as it emerges from the ribosome (21, 31). In the second step, the resulting complex then binds to the SR (32, 33). The primary binding site for the signal sequence in SRP54 is believed to be the deep hydrophobic groove in the M domain, which is bounded on one side by the finger loop (10–12, 28). However, it was shown that the NG domain makes important contributions to the binding process (13, 34–36). This observation indicated a close proximity of the NG domain and the hydrophobic groove in SRP in the targeting mode, and direct NG–M domain communications were supported by the cryo-EM structure of mammalian SRP bound to ribosome and signal sequence (21–23). In the cryo-EM structure, the N domain is anchored on the ribosome, whereas the G domain is separated from the ribosome and SRP RNA but is close to the finger loop (21–23). Consequently, the NG domain is able to sense the loading status of the hydrophobic groove and can adjust the N–G interface accordingly (14, 15, 37). The SRP54 domain arrangement on the ribosome is strikingly different than that observed in S domain A. Here, the SRP54 domains are scaffolded by the SRP RNA so that the NG and the M domains are spatially well separated. In this arrangement, the direct NG–M communications that are thought to be critical for the binding of signal sequences and subsequent signal transduction are unlikely to occur.
It is possible that the SRP RNA, by bridging NG and M domains as seen in S domain A, communicates signal sequence binding to the NG domain, thus stimulating the required domain rearrangement. However, the cryo-EM structure of the SRP bound to an empty ribosome shows that the NG domain is anchored on the ribosome but that the 4.5S RNA/M domain complex is flexible, all of which indicates that the NG domain is separated from the RNA before signal sequence binding (23). Biochemical studies show that the SRP interacts with ribosomes initially only at contacts between the tip of the SRP54 N domain and the ribosomal proteins L23a and L35 (L23 in E. coli) (23, 38–40). Interestingly, in S domain A, the tip of the N domain is involved in interactions with both the RNA and the GM-linker. Hence, a mutually exclusive interaction of the N domain with the 7S RNA/GM-linker and the ribosome may effectively link ribosome interaction with the open S domain structure, thus providing flexibility for scanning ribosomes for the presence of signal sequences.
It has been shown that SRP RNA participates in the central step of protein targeting by catalyzing the interaction of SRP with SR and stimulating the GTPase hydrolysis in the SRP–SR complex once formed (41, 42). Both these RNA activities may be modulated by the presence of the signal sequence and the ribosome (19, 41–45). A recent model of the E. coli SRP–FtsY complex suggests that the SRP RNA localizes at the Ffh–FtsY heterodimer interface on SRP–FtsY association (19). Modeling of FtsY on SRP54 in S domains A and B does not position the RNA close to FtsY. This suggests distinct SRP54 NG–RNA interactions in the free and the SR-bound forms of SRP. Furthermore, it agrees with the suggested rearrangement in the SRP core on SR binding that structurally links the distal helix 8 with its associated M domain to the GTPase centers of SRP and SR (19, 45).
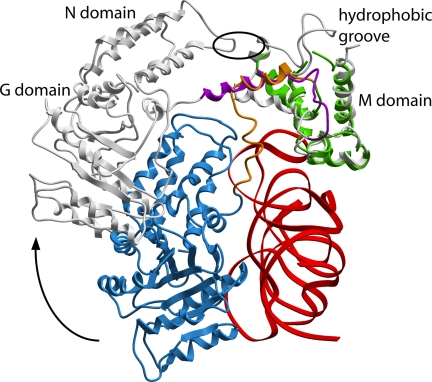
The GM-linker constitutes the key structural element that allows the reported large domain rearrangements within SRP54 linking binding of external ligands by SRP to the acquisition of proper NG–M configurations (8, 16, 19–21). In the open structure of the S. solfataricus SRP core, this GM-linker forms a well defined long α-helix that places the N domain in contact with the fingerloop in the M domain (20). In the crystal of the M. jannaschii SRP core, the GM-linker has two conformations. When the NG domain is RNA-bound, the linker cannot form a long α-helix but bends sharply and contacts both the N domain tip and the RNA. The linker structure in S domain B suggests that the “opening” of SRP is coupled to an unbending and extension of the GM-linker helix. We therefore speculate that the GM-linker might act as a flexible spring between the M and NG domains, to allow for the large ribosome-induced conformational rearrangement within the SRP core (Fig. 5). The hinge regions at the linker N and C termini may then allow further adjustment of the NG–M interface upon signal sequence binding (20).
Fig. 5.
Superposition of the SRP54 M domains of the S. solfataricus SRP54–helix 8 complex (gray) and the M. jannaschii S domain A complex. Shown is the shift in position of the NG domain and the formation of the long GM-linker helix (magenta) when the NG domain is released from the RNA. The color code in the M. jannaschii S domain is as in Fig. 1. SRP19 is omitted for clarity. The position of the hydrophobic contact between the apical loop in the N domain and the finger loop in the M domain in the S. solfataricus SRP54–helix 8 structure is indicated by an oval.
In conclusion, the structure of the M. jannaschii SRP54–SRP19–7S.S RNA complex provides additional evidence for the pivotal role SRP RNA plays in the regulation of the SRP54 activities allowing for an ordered sequence of events during protein targeting.
Methods
Preparation of SRP54–SRP19–RNA Complex.
The 96-nt RNA fragment corresponding to nucleotides G142–G237 of M. jannaschii 7S RNA was transcribed in vitro, and RNA and the SRP19 protein were purified as described in ref. 27. The srp54 gene was PCR amplified from M. jannaschii genomic DNA, and the sequence coding for Met-1–Leu-427 was cloned into the pNZ8048 vector (Leu-2 was omitted to introduce an NcoI restriction site). SRP54 was expressed in Lactococcus lactis with a C-terminal hexahistidine affinity tag, purified by chromatography on Ni-nitrilotriacetic acid, Mono S, heparin Sepharose, and Superdex 75 columns (GE Healthcare, Piscataway, NJ), and stored at −80°C. Before SRP54–SRP19–7S.S RNA complex formation, the RNA was annealed in water by denaturation at 80°C followed by snap cooling on ice. The annealed RNA was purified on Mono Q (GE Healthcare) and dialyzed against 10 mM Tris·HCl (pH 7.5)/5 mM MgCl2. The complex was reconstituted in a buffer containing 10 mM Tris·HCl (pH 8.0), 250 mM KCl, 5 mM MgCl2, 1% β-mercaptoethanol, and 5% glycerol. Binding reactions were incubated for 15 min at room temperature after addition of SRP19, and for 1 h at 37°C after addition of SRP54. After this, the final purification of the complex was performed on Mono Q. The purified complex behaves as a monomer in size exclusion chromatography (90 kDa; data not shown).
Crystallization, Data Collection, and Structure Determination.
The SRP54–SRP19–7S.S RNA complex (3 mg/ml) was crystallized by the hanging-drop vapor-diffusion technique at 18°C. Crystals grew in 5 days when the protein solution was mixed with an equal volume of mother liquor containing 8% polyethylene glycol 6000, 100 mM Mes (pH 5.4), 50 mM Na-Citrate, 30 mM MgCl2, and 3% dimethylsulfoxide, and equilibrated against the same solution. Crystals (0.4 × 0.4 × 0.1 mm3) were cryocooled and diffraction data were measured on a marMosaic 225 CCD detector at the European Synchrotron Radiation Facility ID23-1 beamline by using x-ray radiation with λ = 0.979 Å at 100 K. Diffraction data from one crystal were reduced and scaled by using the CCP4 Suite (46). The crystals belong to space group P212121 with cell parameters a = 70.29 Å, b = 128.40 Å, c = 163.42 Å, with two S domains in the asymmetric unit and a solvent content of 40.1%.
The structures of the M. jannaschii SRP19–7S.S complex [Protein Data Bank (PDB) ID code 1LNG (27)] and the E. coli M domain [PDB ID code 1DUL (28)] and x-ray data from 11.0- to 2.7-Å resolution were used in molecular replacement searches with the programs CNS (47) and MOLREP (46). FFFEAR (46) was used for detection of secondary structural elements in the electron density maps. The NG domains were modeled from Thermus aquaticus (PDB ID code 2FFH), manually docked into the electron density. The model was built in O (48) and refined by CNS and REFMAC (46) by using the maximum-likelihood residual, anisotropic scaling, bulk-solvent correction, and atomic displacement parameter refinement as used in the translation, libration, screw-rotation (TLS) method (49) against all data from spacings between 20 and 2.5 Å. The choice of TLS groups was made by exploring the significance of the improvement made to the refinement as the structure was progressively divided into smaller TLS groups of atoms. The TLS parameters for the NG domain in S domain B refined to larger values, reflecting the larger overall displacements of this domain. The overall improved description of the structure was reflected in the Rwork and Rfree values, which fell by 2.3 and 2.7%, respectively. Noncrystallographic symmetry restraints were used at the beginning, but not at the end of the refinement. The Rwork and Rfree for the final model are 24.4 and 29.4%, respectively. The GM-linker, Leu-299–Arg-323, could be traced in both S domains A and B, even though the electron density is weak. The kink at Asp-311 in S domain A, which is missing in S domain B, is clearly defined. Data collection and refinement statistics are summarized in Table 1. Figs. 1 B and C, 2A, 4, and 5, and SI Figs. 6, 8, and 10 were prepared with ICM (50); Figs. 2B and 3B were prepared with MOLMOL (51); and Fig. 3A was prepared with PYMOL (52).
Supplementary Material
Acknowledgments
We thank the European Synchrotron Radiation Facility (Grenoble, France) and the MAX Laboratory (Lund, Sweden) for provision of synchrotron radiation facilities; Edward Mitchell (beamline ID23-1, European Synchrotron Radiation Facility) for excellent assistance; and T. Bergfors for critical reading of the manuscript. This work was supported by Swedish Research Council Grant 621-2002-5723, European Union Grant CLK3-CT-2000-00082, and the Gustafsson Foundation.
Abbreviations
- SRP
signal-recognition particle
- TLS
translation, libration, and screw rotation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors for the Methanococcus jannaschii SRP54–SRP19–RNA complex structure have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2v3c).
This article contains supporting information online at www.pnas.org/cgi/content/full/0702467104/DC1.
References
- 1.Doudna JA, Batey RT. Annu Rev Biochem. 2004;73:539–557. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- 2.Luirink J, Sinning I. Biochim Biophys Acta. 2004;1694:17–35. doi: 10.1016/j.bbamcr.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 3.Egea PF, Stroud RM, Walter P. Curr Opin Struct Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Walter P, Blobel G. J Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen ES, Rosenblad MA, Larsen N, Westergaard JC, Burks J, Wower IK, Wower J, Gorodkin J, Samuelsson T, Zwieb C. Nucleic Acids Res. 2006;34:D163–D168. doi: 10.1093/nar/gkj142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Römisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein HD, Poritz MA, Strub K, Hoben PJ, Brenner S, Walter P. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- 8.Freymann DM, Keenan RJ, Stroud RM, Walter P. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 9.Montoya G, Kaat K, Moll R, Schafer G, Sinning I. Structure (London) 2000;8:515–525. doi: 10.1016/s0969-2126(00)00131-3. [DOI] [PubMed] [Google Scholar]
- 10.Keenan RJ, Freymann DM, Walter P, Stroud RM. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 11.Zopf D, Bernstein HD, Johnson AE, Walter P. EMBO J. 1990;9:4511–4517. doi: 10.1002/j.1460-2075.1990.tb07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.High S, Dobberstein B. J Cell Biol. 1991;113:229–233. doi: 10.1083/jcb.113.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lütcke H, High S, Romisch K, Ashford AJ, Dobberstein B. EMBO J. 1992;11:1543–1551. doi: 10.1002/j.1460-2075.1992.tb05199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 15.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu F, Shan SO, Moustakas DT, Alber F, Egea PF, Stroud RM, Walter P, Burlingame AL. Proc Natl Acad Sci USA. 2004;101:16454–16459. doi: 10.1073/pnas.0407456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buskiewicz I, Kubarenko A, Peske F, Rodnina MV, Wintermeyer W. RNA. 2005;11:947–957. doi: 10.1261/rna.7242305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buskiewicz I, Peske F, Wieden HJ, Gryczynski I, Rodnina MV, Wintermeyer W. J Mol Biol. 2005;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 19.Spanggord RJ, Siu F, Ke A, Doudna JA. Nat Struct Mol Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- 20.Rosendal KR, Wild K, Montoya G, Sinning I. Proc Natl Acad Sci USA. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 22.Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Nature. 2006;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 23.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 24.Lentzen G, Moine H, Ehresmann C, Ehresmann B, Wintermeyer W. RNA. 1996;2:244–253. [PMC free article] [PubMed] [Google Scholar]
- 25.Diener JL, Wilson C. Biochemistry. 2000;39:12862–12874. doi: 10.1021/bi001180s. [DOI] [PubMed] [Google Scholar]
- 26.Gu SQ, Jockel J, Beinker P, Warnecke J, Semenkov YP, Rodnina MV, Wintermeyer W. RNA. 2005;11:1374–1384. doi: 10.1261/rna.7219805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hainzl T, Huang S, Sauer-Eriksson AE. Nature. 2002;417:767–771. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- 28.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 29.Kuglstatter A, Oubridge C, Nagai K. Nat Struct Biol. 2002;9:740–744. doi: 10.1038/nsb843. [DOI] [PubMed] [Google Scholar]
- 30.Hainzl T, Huang S, Sauer-Eriksson AE. RNA. 2005;11:1043–1050. doi: 10.1261/rna.2080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter P, Ibrahimi I, Blobel G. J Cell Biol. 1981;91:545–550. doi: 10.1083/jcb.91.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilmore R, Blobel G, Walter P. J Cell Biol. 1982;95:463–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilmore R, Walter P, Blobel G. J Cell Biol. 1982;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zopf D, Bernstein HD, Walter P. J Cell Biol. 1993;120:1113–1121. doi: 10.1083/jcb.120.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newitt JA, Bernstein HD. Eur J Biochem. 1997;245:720–729. doi: 10.1111/j.1432-1033.1997.00720.x. [DOI] [PubMed] [Google Scholar]
- 36.Cleverley RM, Gierasch LM. J Biol Chem. 2002;277:46763–46768. doi: 10.1074/jbc.M207427200. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez UD, Minasov G, Focia PJ, Stroud RM, Walter P, Kuhn P, Freymann DM. J Mol Biol. 2002;320:783–799. doi: 10.1016/s0022-2836(02)00476-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 39.Gu SQ, Peske F, Wieden HJ, Rodnina MV, Wintermeyer W. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ullers RS, Houben EN, Raine A, ten Hagen-Jongman CM, Ehrenberg M, Brunner J, Oudega B, Harms N, Luirink J. J Cell Biol. 2003;161:679–684. doi: 10.1083/jcb.200302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peluso P, Herschlag D, Nock S, Freymann DM, Johnson AE, Walter P. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- 42.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 43.Bacher G, Lutcke H, Jungnickel B, Rapoport TA, Dobberstein B. Nature. 1996;381:248–251. doi: 10.1038/381248a0. [DOI] [PubMed] [Google Scholar]
- 44.Bradshaw N, Walter P. Mol Biol Cell. 2007;18:2728–2734. doi: 10.1091/mbc.E07-02-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Siu FY, Spanggord RJ, Doudna JA. RNA. 2007;13:240–250. doi: 10.1261/rna.135407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collaborative Computational Project N. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 47.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 48.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 49.Winn MD, Isupov MN, Murshudov GN. Acta Crystallogr D. 2001;57:122–133. doi: 10.1107/s0907444900014736. [DOI] [PubMed] [Google Scholar]
- 50.Abagyan R, Totrov M. J Mol Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- 51.Koradi R, Billeter M, Wuthrich K. J Mol Graphics. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 52.DeLano WL. The PyMOL Molecular Graphic System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.