Fig. 1.
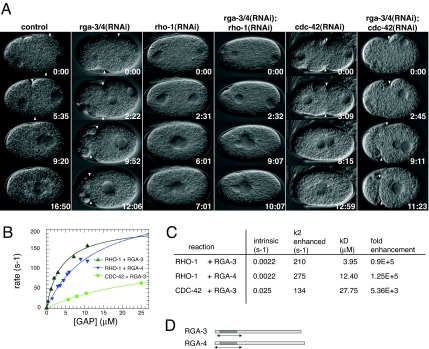
RGA-3 and RGA-4 are GAPs for RHO-1. (A) Time-lapse differential interference contrast images of control and RNAi embryos during polarity establishment. In this and subsequent figures the embryos are ≈50 μm long. Embryo posterior is to the right. In rga-3/4(RNAi) embryos, the cortex is hypercontractile and the boundary between the contractile anterior cortex and the noncontractile posterior cortex (PCF, white arrowhead) is shifted toward the anterior. The relaxation of the PCF is delayed in rga-3/4(RNAi) embryos. In rho-1(RNAi) and rga-3/4(RNAi);rho-1(RNAi) embryos cortical contractility is abolished, whereas in rga-3/4(RNAi);cdc-42(RNAi) embryos the contractility resembles rga-3/4(RNAi) embryos. Times (minutes:seconds) are standardized to similar position of the pronuclei at the beginning of pronuclear migration. (B) Observed hydrolysis rates for 100 nM RHO-1 or CDC-42 in the presence of the indicated amounts of RGA-3 (green) or RGA-4 (blue). Solid lines represent hyperbolic fits to the data points, from which the values in C were extracted. (C) Comparison between intrinsic RHO-1/CDC-42 GTP hydrolysis rates and maximally attainable ones in the presence of RGA-3 or RGA-4 GAP domains. The binding constant (affinity) between the GTPase and the GAP is represented by kD. Fold enhancement is the ratio between the intrinsic and the maximally attainable, stimulated rate. (D) Predicted GAP domains (dark shading) and the soluble GAP domains used in this analysis (arrows).