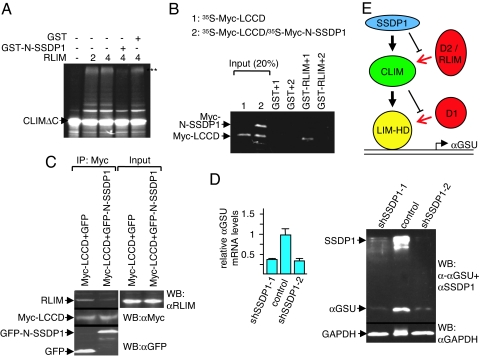
Fig. 4.
SSDP1 inhibits binding of RLIM to CLIM and regulates Lhx3 target gene expression. (A) In vitro ubiquitination experiment using 35S-labeled CLIMΔC, RLIM as E3, and UbcH5 as E2 enzyme in the presence of GST-N-SSDP1 or GST alone. Arrows point at unmodified CLIM proteins. Polyubiquitinated proteins are indicated by three asterisks (***). Note the partial inhibition of CLIMΔC ubiquitination by N-SSDP1. (B) N-SSDP1 inhibits binding of RLIM to the LCCD. Shown is the GST pull-down using GST-RLIM and 35S-Myc-LCCD (1) or Myc-LCCD cotranslated with Myc-N-SSDP1 (35S-Myc-LCCD/35S-Myc-N-SSDP1) (2). (C) Co-IP of endogenous RLIM in cells cotransfected with Myc-LCCD and GFP-N-SSDP1. Note the decreased RLIM precipitation in the presence of N-SSDP1. (D) Down-regulation of SSDP1 results in lower levels of αGSU mRNA and protein. (Left) SSDP1 levels were knocked-down in αT3 cells via retroviral infection of mouse shRNAs (shSSDP1-1, -2) or the empty retroviral vector. mRNA encoding αGSU was measured by qRT-PCR (n = 3; values are mean ± SE). (Right) Western blot of the same shRNA-treated cells. Note that knocking down endogenous SSDP1 levels leads to a significant decrease in endogenous αGSU levels at the mRNA and protein levels. (E) Shown is a model of a cascade of protein interactions that protects LIM-HDs from proteasomal degradation. Via binding to LIM domains, a destabilizing enzyme (D1, red) targets LIM-HDs (yellow) for degradation. In the presence of CLIM (green), the binding of D1 to LIM domains is inhibited, resulting in stabilization of LIM-HDs. CLIM is targeted by another destabilizing enzyme(s) (D2/RLIM) for ubiquitination/degradation. The presence of SSDP1 (blue) prevents binding of D2/RLIM to CLIM thereby protecting the LIM-HD/CLIM protein complex.