Abstract
Arabidopsis thaliana histone deacetylase 1 (AtHD1 or AtHDA19), a homolog of yeast RPD3, is a global regulator of many physiological and developmental processes in plants. In spite of the genetic evidence for a role of AtHD1 in plant gene regulation and development, the biochemical and cellular properties of AtHD1 are poorly understood. Here we report cellular localization patterns of AtHD1 in vivo and histone deacetylase activity in vitro. The transient and stable expression of a green fluorescent (GFP)-tagged AtHD1 in onion cells and in roots, seeds and leaves of the transgenic Arabidopsis, respectively, revealed that AtHD1 is localized in the nucleus presumably in the euchromatic regions and excluded from the nucleolus. The localization patterns of AtHD1 are different from those of AtHD2 and AtHDA6 that are involved in nucleolus formation and silencing of transgenes and repeated DNA elements, respectively. In addition, a histone deacetylase activity assay showed that the recombinant AtHD1 produced in bacteria demonstrated a specific histone deacetylase activity in vitro. The data suggest that AtHD1 is a nuclear protein and possesses histone deacetylase activities responsible for global transcriptional regulation important to plant growth and development.
Keywords: Arabidopsis, histone deacetylase, RPD3, subcellular localization, gene silencing, chromatin
Introduction
The reversible acetylation and deacetylation of specific lysine residues on core histone N-terminal tails is catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. The competing action between HATs and HDACs determines histone acetylation levels that ultimately affect higher-order chromatin structure and gene activities [1-3]. In general, hyperacetylated histones are associated with gene activation, whereas hypoacetylated histones are related to gene repression. After the first histone deacetylase (HDAC1), a yeast RPD3 homologue, was of HDACs have been characterized in many eukaryotes, and 18 HDAC-like genes were predicted in Arabidopsis [5]. HDACs can be divided into 4 classes based on their sequence homology, substrate specificity, and cofactor requirement [6, 7]. Class I and class II HDACs are homologous to the yeast RPD3 and HDA1 proteins, respectively, while class III HDACs are related to yeast SIR2 protein. The fourth class of HDACs, known as HD2, is found only in plants [8].
In yeast and mammalian cells, the Rpd3-like HDACs mediate transcriptional repression by interacting with specific DNA-binding proteins or associated with corepressors and by recruiting to target promoters [9-11]. RPD3 and Sin3 homologues are associated with a large multiprotein complex that represses target genes involved in diverse processes such as meiosis, cell-type specificity, potassium transport, phosphate and phospholipid metabolism, methionine biosynthesis, and responses to stress and developmental changes [12-18].
In plants, the first RPD3 homolog was identified in maize, which complemented the phenotype of an rpd3 null mutant of yeast [19]. Arabidopsis has four RPD3-like HDACs including AtHD1 (AtRPD3A, HDA19), AtHD6 (AtRPD3B), HDA7 and HDA9 [5], each of which might have evolved to possess specific functions. For instance, HDA6 was associated with transgene silencing, DNA methylation, and regulation of rRNA genes [20-23].
Down-regulation of AtHD1 by antisense inhibition and T-DNA insertion in Arabidopsis led to various phenotypic and developmental defects including early senescence, serrated leaves, formation of aerial rosettes, delayed flowering, and defects in floral organ identity [24, 25]. Further by DNA microarray showed that the down-regulation of AtHD1 induced up- and down-regulation of >7% transcriptome including the genes involved in cellular biogenesis, protein synthesis, ionic homeostasis, protein transcription, and plant hormonal regulation [26]. The data suggest that AtHD1 is a global transcriptional regulator that provides reversible gene regulation in response to changes in developmental programs and physiological processes. AtHD1 may also play a role in plant defense as it can be up-regulated by wounding, infection of a pathogenic fungus, and plant hormones induced in pathogen response [27].
The histone deacetylase activity of AtHD1 is implied by the concomitant changes in histone acetylation and AtHD1 expression levels. The up- and down-regulation of AtHD1 was associated with the hypoacetylation and hyperacetylation, respectively, of histone H4 in transgenic Arabidopsis [25-27]. Here, we report histone deacetylase activity of recombinant AtHD1 in vitro. Subcellular localization of AtHD1 suggests that AtHD1 is a nuclear protein and does not interact with nucleolus, suggesting a specific role of AtHD1 in global gene regulation that is different from that of AtHD2 or AtHDA6 and other AtHD1-related homologs.
Materials and Methods
Plant materials
Seeds of Arabidopsis thaliana ecotypes (Col, Ler, and Ws) were obtained from Arabidopsis Biological Resources Center (ABRC) and sown directly in vermiculite mixed with 10% soil. Seeds from transgenic Arabidopsis were sterilized in 10% sodium hypochlorite (commercial bleach), rinsed three times in sterilized water, and placed for germination in Murasige/Skoog (MS) medium (Sigma, St. Louis) containing 50 mg/L kanamycin [25]. After two weeks, the seedlings were transferred to soil mix and grown in growth chamber. All plants were grown in a growth chamber with a cycle of 14 h of light at 22 °C and 10 h of dark at 18 °C.
Construction of 35S∷AtHD1-GFP chimeric gene
The AtHD1 full-length cDNA (AF014824) was amplified from a plasmid clone pAtHD1-2 (Tian and Chen, 2001) by PCR using primer pair 5′-GCG TCG AC A TGG ATA CTG GGG GCA ATT C-3′ (forward) and 5′-CAT GCC ATG GCT GTT TTA GGA GGA AAC GCC TG-3′ (reverse). The resulting PCR product was digested with SalI and NcoI and subcloned into the plasmid pUC18/CaMV35S-GFP(S65T) so that the 3′-end of AtHD1 coding sequence was fused to the 5′-end of GFP in frame, and the whole AtHD1-GFP chimeric gene was under the controlofthecauliflowermosaic virus (CaMV) 35S promoter. The expression cassette of 35S∷AtHD1-GFP was excised from the pUC18 plasmid by XbaI and EcoRI double digestion and subcloned into the binary vector pBI101 for plant transformation. A 35S∷GFP construct was used as control for localization studies.
DNA and RNA blot analyses
Genomic DNA (2 μg) from wild type and SHG transgenic plants were digested with EcoRI, separated by electrophoresis in a 1% analysis agarose gel, and transferred to Hybond-N+ membrane (Amersham Pharmacia). A radiolabeledprobe(0.7-kb)specific for the GFP sequence was used in hybridization (Figure 1A) by a random priming method (Amersham). The plasmid pUC18/35S∷AtHD1-GFP digested with EcoRI was used as a positive control (lane V). DNA markers (M) used were Lamda DNA digested with EcoRI and HindIII (Promega).
Figure 1.
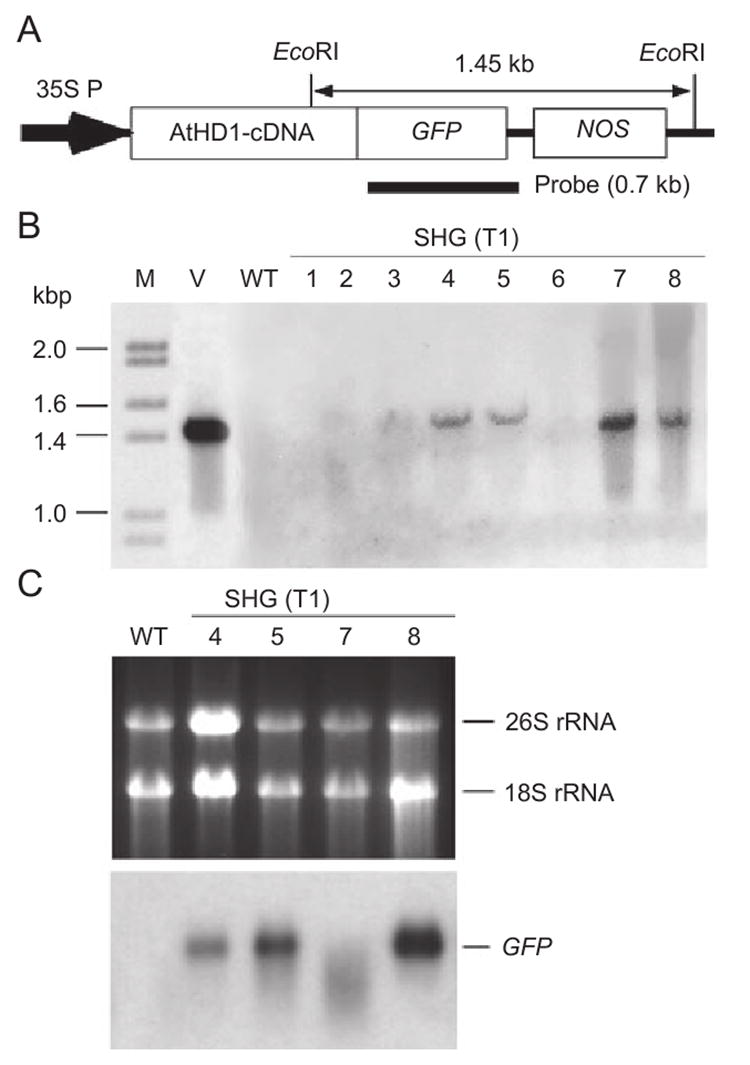
(A) The diagram of AtHD1-GFP chimeric gene cassette. 35SP: CaMV 35S promoter and NOS Ter: nopaline synthase terminator (NOS ter). Sizes of the AtHD1-GFP chimeric gene and recombinant protein are 2.2 kb and 83.1 kDa, respectively. The genomic fragment containing the transgene and the probe used for DNA and RNA blot analyses are shown above and below the diagram, respectively. (B) Genomic DNAs from a control plant and eight transgenic plants were digested with EcoRI, blotted, and hybridized with the probe indicated in Figure 1A. The transgene (a 1.45 kb fragment containing the GFP coding sequence) was detected in four of eight SHG T1 lines examined. (C) Northern blot analysis using the radiolabeled probe specific for the GFP sequence showed that the AtHD1-GFP transcript was present in three of four SHG T1 transgenic lines examined. The ethidium bromide-stained RNA gel depicting the rRNAs is shown as loading controls.
Total RNA (20 μg) from wild type and SHG transgenic plants (with the presence of transgene separated by electrophoresis in a 1.5% agarose gel containing 2% formaldehyde and then transferred to a Hybond-N+ membrane (Amersham Pharmacia). The membrane was hybridized with a radio-labeled probe (Figure 1A), and the signals were detected as previously described [25].
Transient expression assays
The pUC18/35S∷AtHD1-GFP and 35S∷GFP plasmids were introduced into onion cells by particle bombardment. Fresh onion bulbs purchased from local grocery store were cut into slices of 1 cm2 and placed on the central area of a petri dish with 0.9% phytal agar as supporting medium. Tungsten particles of 1.1 mm in diameter were prepared and coated with plasmid DNA according to the manufacturer’s recommendations (BioRad). Bombardments were performed using the Biolistic PDS-1000/He Particle Delivery System (BioRad) with the following parameters: holder level, 2; sample level, 4; gap distance, 0.63 cm; target distance, 6 cm; helium pressure, 1100 psi; chamber vacuum, 27 inches Hg (~ 0.06 atm); amount of DNA/bombardment, 1mg; and amount of macrocarrier per bombardment, 500 mg of tungsten particles. Each sample received two times of bombardment, after which the samples were incubated in a growth chamber at 25 °C for 20–24 h. A single layer of epidermal cells was peeled from the onion scale leaves and examined for AtHD1-GFP expression under a fluorescence microscope.
Arabidopsis transformation and selection
Ten Arabidopsis thaliana (Columbia) plants were transformed with Agrobacteria harboring the binary vector pBI101/35S∷AtHD1-GFP using the floral dip method [28]. Seeds of T0 plants were selected by germination on MS medium (Sigma) with 50 mg/L kanamycin. Successful transformants (T1 plants) were regenerated and grown in soil. Transformation efficiency is about 1%. The presence of AtHD1-GFP transgene was detected in the T1 plants by PCR using the following primers flanking the GFP coding sequence: 5′-ATG GTG AGC AAG GGC GAG-3′ (forward) and 5′-TTA CTT GTA CAG CTC GTC CA-3′ (reverse).
Detection of GFP signals
Transient expression of AtHD1-GFP fusion protein in onion epidermal cells was detected with the Olympus AX7 fluorescence microscope. A single epidermal layer of the transformed onion scale leaf disc was peeled off and put on a slide for examination under 60 × or 100 × magnifications. Nuclei were stained by adding one drop of 4′,6-diamidino-2-phenylindole (DAPI) staining solution (1 μg/ml DAPI in PBS) to the onion cells, and DAPI fluorescence images were obtained by UV illumination of the specimen and a band-pass filter for DAPI. Green fluorescence signals were obtained by blue light illumination and a band-pass filter for GFP. Stable expression of AtHD1-GFP fusion protein in transgenic Arabidopsis was detected by confocal scanning microscopy. Dissected intact tissues were stained with DAPI staining solution and examined by differential interference contrast (DIC) and fluorescence microscopy sequentially using the Zeiss Axiophot microscope with 40 × or 100 × magnifications. Images of tissues at different confocal layers (Z-series) were taken with a 0.5 mm step size. GFP fluorescence images were acquired using a GFP filter set (Chroma Technologies, excitation HQ470/20, dichroic Q495LP, emission HQ525/50) and a Photometrics Coolsnap digital camera.
Production of recombinant AtHD1 and western blot analysis
The AtHD1 full-length cDNA was subcloned in frame into the pET21a expression vector (Novagen), which was then transformed into Escherichia coli (strain BL21). Recombinant AtHD1 production in bacteria was induced by isopropyl-β-D-thiogalactoside. Total protein was extracted from bacterial cells by sonication, and recombinant AtHD1 protein was purified by Ni-NTA affinity column (Invitrogen) followed by amylose affinity column (New England Biolabs). Affinity tag (6×His) was subsequently removed by tobacco etch virus protease cleavage. Recombinant AtHD1 protein was separated by 15% SDS-PAGE and transferred to PVDF membrane (Invitrogen) using a Trans-Blot SD Semi-dry Transfer Cell (Bio-Rad). Western blot analysis was performed according to the protocol of Western Breeze kit (Invitrogen). The primary antibody was anti-HD1 as previously described in Tian and Chen (2001). The secondary antibody used was an anti-rabbit antibody conjugated with alkaline phosphatase. Detection was carried out by adding a chemiluminescent substrate (CPD-star) to the PVDF membrane, and the signals were detected with KodakX-rayfilm. AtHDA6, AtHDA9, and AtHDA17 were also produced using the same procedure in E. coli cells and baculovirus expression system in SF9 insect cells (Invitrogen Inc.). The data were not shown because none of other proteins showed any measurable levels of histone deacetylase activity.
Histone deacetylase activity assays
Synthetic histone H3 and H4 peptides were acetylated and labeled according to the protocol of the HDAC Activity Assay kit (Upstate Biotechnology). Recombinant AtHD1 protein was added to the same amount of tritium-labeled acetylated histone H3 or H4 peptides to test for HDAC activity. For each test sample, two sets of duplicate assays with 50 mM sodium butyrate (SB), a specific inhibitor of HDAC, were includedtodemonstratethespecificityof deacetylation. HeLa nuclear extract that contains a blend of histone deacetylases was used as the positive control. The negative control was water (data not shown) and water plus SB. After 16–20 h of incubation at room temperature, the reaction mixtures were centrifuged and the radioactivity of the supernatant was measured by scintillation counting using the Beckman LS 6500 Scintillation counter.
Results
Expression of AtHD1-GFP fusion protein in transgenic Arabidopsis
A chimeric gene consisting of an engineered green fluorescent protein (GFP) sequence was fused to the C-terminus of AtHD1 cDNA (Figure 1A). Transgenic Arabidopsis plants carrying the same AtHD1-GFP chimeric gene were generated by Agrobacterium-mediated transformation [29, 30]. These plants were named “SHG” for the transgene (35S∷AtHD1-GFP) they contain. In the first round of selection (T0), the transformation efficiency was about 1%, which is a typical value for the floral dipping method. The panels), seeds collected from the firstgeneration of transformants (T0) were germinated in selective medium to produce T1 transgenic plants, which were used for the assay of AtHD1-GFP gene expression.
The results of Southern blot analysis (Figure 1B) show that the 35S∷AtHD1-GFP transgene was present in four of the eight transgenic lines examined. The same probe used in Southern blot analysis was used in RNA blot analysis. The AtHD1-GFP transcripts were detected in three of four transgenic lines examined (Figure 1C). SHG7 may contain the transgene (lane 7, Figure 1B) but the RNA was degraded (Figure 1C). Alternatively, the transgene is silenced in this line. SHG lines 5 and 8 were selected for AtHD1 cellular localization assays.
Subcellular localization of AtHD1
To study the subcellular localization of AtHD1, the chimeric gene (Figure 1A) was expressed transiently in onion cells and stably in Arabidopsis. In the transient expression assay, the 35S∷AtHD1-GFP chimeric gene was introduced into onion epidermal cells by particle bombardments. The cells were stained with DAPI (4′,6-diamidino-2-phenylindole) to reveal the nuclei and then examined using a fluorescence microscope. GFP signals were detected in the nuclei of onion cells (Figure 2, second panel), whereas the control (35S∷GFP) displayed diffused patterns of localization (data not shown). The AtHD1-GFP chimeric gene was introduced into Arabidopsis plant genome by Agrobacterium-mediated transformation, and the transgenic plants were named SHG for the transgene (35S∷AtHD1-GFP) they contained. Different tissues of the SHG transgenic plants were dissected, stained with DAPI, and then examined by confocal fluorescence microscopy. In all tissues examined, including roots, leaves, flowersand developing seeds in the siliques (Figure 2, third-fifth signals were detected and localized in the nuclei, which is consistent with recent findings [27]. We further analyzed AtHD1 localization patterns using high-resolution confocal image analysis. We found that AtHD1-GFP protein appeared to be excluded from the nucleolus because GFP signals were absent in the nucleolus (Figure 3). This is in contrast to the HD2-type HDACs that are localized in the nucleolus in maize [8] and Arabidopsis [31]. Moreover, the localization pattern of AtHD1 is different from that of AtHDA6 that is primarily associated with rDNA and heterochromatic regions [23]. To visualize detailed staining patterns, mesophyll cells at early prophase of the cell cycle (Figure 2Q-2S) are shown in Figure 3E-3G (enlarged scales). The DAPI fluorescence image (Figure 3F) shows that the chromatin began to condense into chromatids, and the nucleolus was obviously formed in the nucleus. The corresponding GFP fluorescence image (Figure 3G) shows that the AtHD1-GFP protein was absent in the nucleolus, and it aggregated into speckles in the nucleoplasm. The AtHD1-GFP protein may be associated with the condensing chromatids, suggesting a role of AtHD1 in mitosis.
Figure 2.
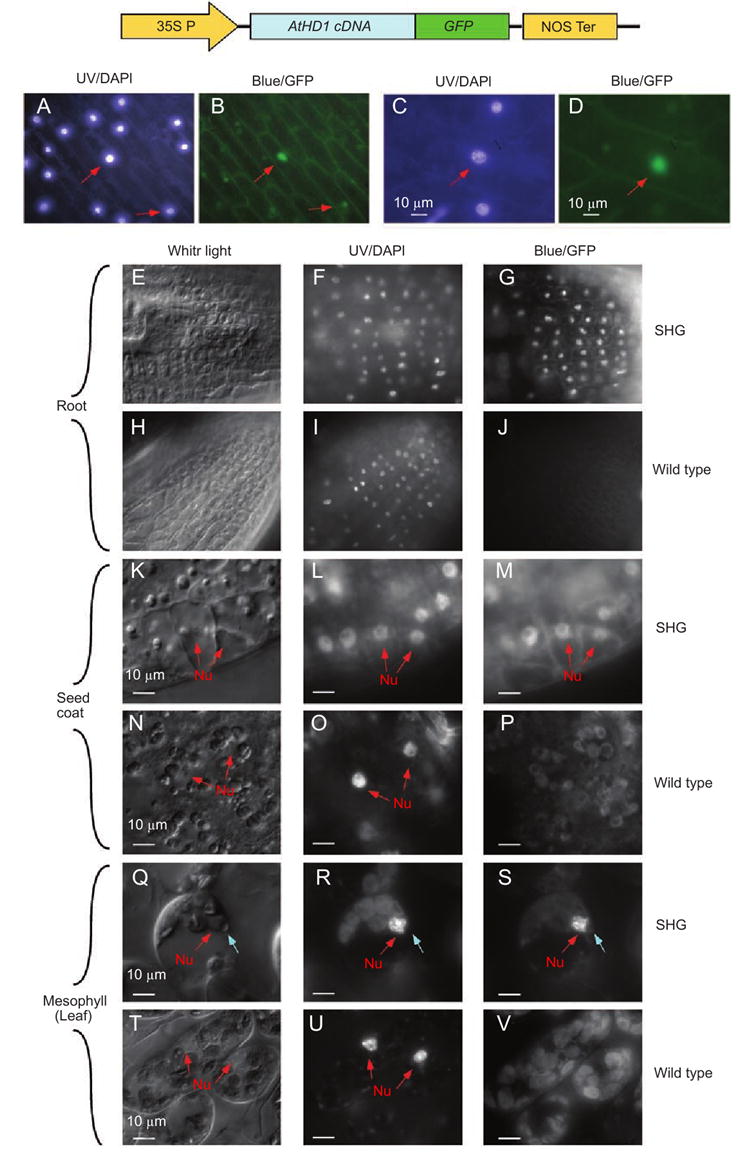
Localization of AtHD1-GFP fusion proteins. Transient expression of AtHD1-GFP fusion protein. (A-D) localization of AtHD1-GFP fusion protein in onion cells. (A) and (C) onion cells were stained with DAPI and viewed under the UV light. The bluish white spots are the nuclei (arrows). (B) and (D), GFP signals (arrows) detected in the same areas as shown in (A) and (C), respectively. Localization of AtHD1-GFP fusion protein in Arabidopsis transgenic plants. (E-J) expression of AtHD1-GFP fusion protein in root epidermal cells. Confocal microscopic images detected by DIC (E and H), DAPI-staining (F and I), and GFP-filter(G and J) in Arabidopsis SHG transgenic (E-G) and WT (H-J) plants, respectively. GFP signals were localized in the nuclei of the root epidermal cells in the SHG plants (G) but not in the WT plant (J). Note the auto-fluorescent interference was very low(J) because of no chloroplasts in the roots. (K-P) expression of AtHD1-GFP fusion protein in developing seed coats. Confocal microscopic images detected by DIC (K and N), DAPI-staining (L and O), and GFP-filter (M and P) in Arabidopsis SHG transgenic (K-M) and WT (N-P) plants, respectively. Nuclei are indicated by arrows. GFP signals were detected in the nuclei of SHG plant cells (M) but not in the WT plants (P). The background was caused by auto-fluorescence of chlorophylls in the chloroplasts. (Q-V), expression of AtHD1-GFP fusion protein in leaf mesophyll cells. Confocal microscopic images detected by DIC (Q and T), DAPI-staining (R and U), and GFP-filter (S and V) in Arabidopsis SHG transgenic (Q-S) and WT (T-V) plants, respectively. GFP signals were detected in the nuclei of the SHG plants (S) but not in the WT plants (V). The nuclei (Nu) and nucleoli (No) were clearly visible in the DIC, DAPI-stained, and GFP-activated image of the SHG plants.
Figure 3.
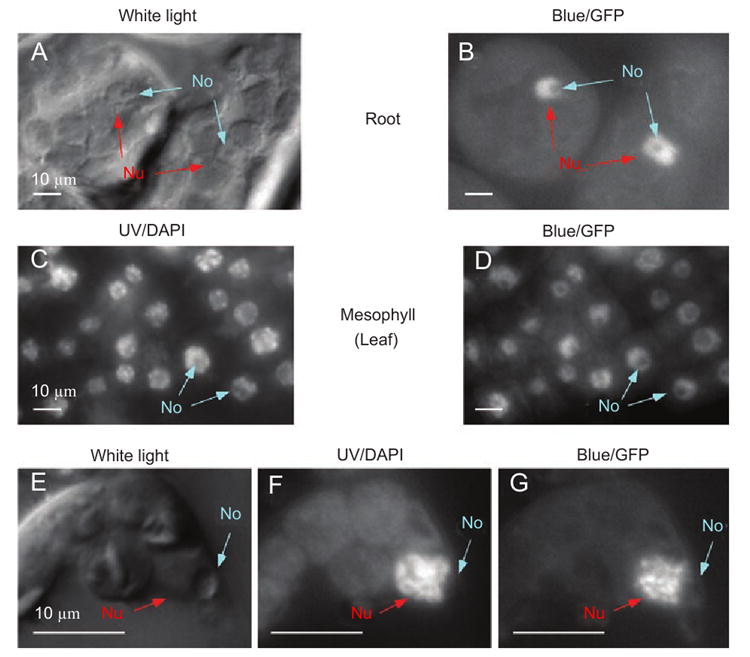
AtHD1-GFP fusion protein is excluded from nucleoli and aggregates during the prophase of cell cycle. (A-B) confocal microscopic images of nuclei (Nu) and nucleoli (No) detected by DIC (A) and GFP-filter (B) in the root cells of SHG transgenic plants. (C-D) confocal microscopic images of nuclei (Nu) and nucleoli (No) detected by DAPI-staining (C) and GFP-filter (D) in the leaf mesophyll cells of SHG transgenic plants. (E-G) enlarged images of nuclei and nucleoli detected by DIC (E), DAPI-staining (F), and GFP-filter (G) in the mesophyll cells of SHG plants (same as in Figure 2 Q-S). (F) DAPI-stained image displayed visible organization of chromatin into chromatids in the prophase of cell division. (G) GFP signals appeared as speckles that were localized in the nucleus but excluded from the nucleolus.
Histone deacetylase activity in the recombinant AtHD1 protein
Recombinant AtHD1 was produced by bacterial expression system for studying histone deacetylase activity. The recombinant protein consists of a hexahistidine tag (6 × His) for protein purification, a maltose binding protein (MBP) for increasing the solubility of the recombinant protein so as to prevent the formation of inclusion bodies in bacterial cells, a tobacco etch virus (TEV) cleavage site for removing the N-terminal segment, and the full-length AtHD1 polypeptide (Figure 4A). Recombinant AtHD1 was purified from the bacterial protein extract by affinity column with Ni-NTA resins specific for 6 × His tag followed by further purification by amylose-conjugated agarose resins for the MBP fragment. The N-terminal segment containing the 6 × His tag and MBP segment was removed from the recombinant protein by TEV protease. The identity of the AtHD1 protein was confirmed by western blot analysis using an antibody specific for the N-terminal region of AtHD1 [25] (Figure 4B).
Figure 4.
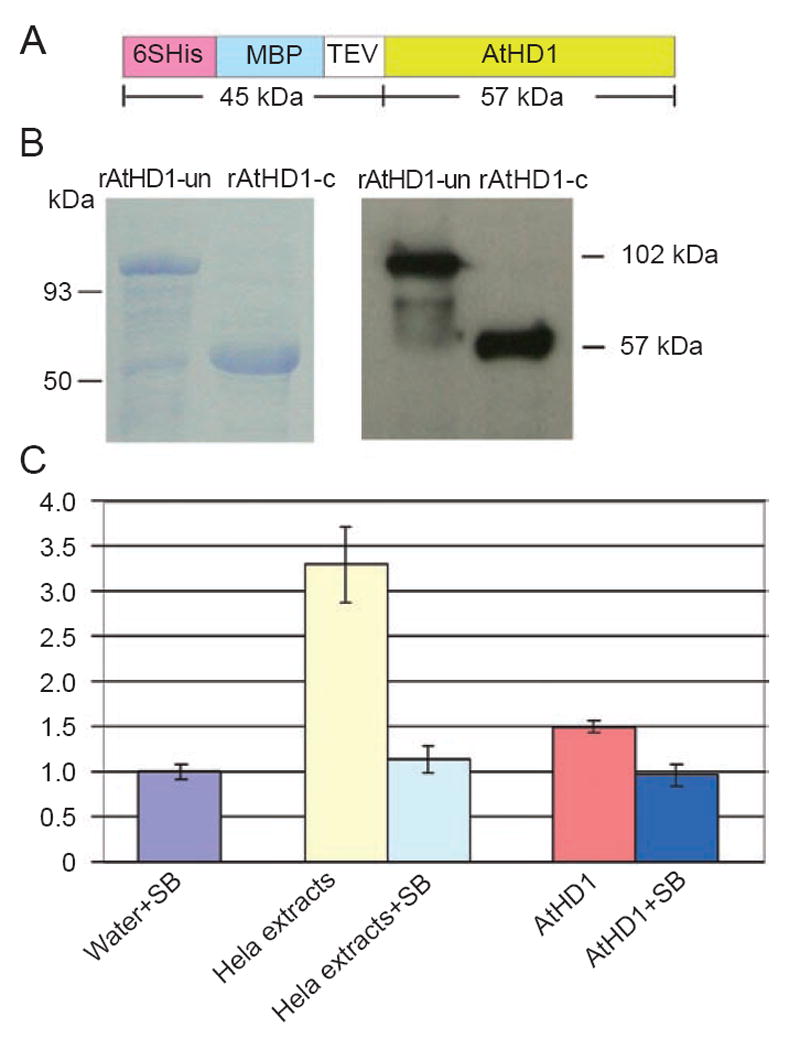
Biochemical assays of recombinant AtHD1. (A) diagram of the expression cassette of recombinant AtHD1 driven by the inducible viral T7 promoter. Upstream AtHD1 there is a hexa-histidine tag (6 × His) for purification by affinity column, a maltose-binding protein (MBP) segment for increasing the solubility of the recombinant protein, and a tobacco etch virus (TEV) protease cleavage site for removal of the 6 × His-MBP segment. Bacteria containing the plasmid encoding recombinant AtHD1 were incubated at 25 °C for 16 h after the addition of IPTG. (B) SDS-PAGE and western blot analyses of recombinant AtHD1. The recombinant AtHD1 protein (rAtHD1) was purified from bacterial crude protein extracts by Ni-NTA affinity column followed by amylose affinity column. The rAtHD1 was subsequently cleaved by TEV protease to remove 6 × His-MBP. The rAtHD1 proteins before and after TEV cleavage had molecular weights of 100 kDa and 57 kDa, respectively, and they were visible as a prominent band in the SDS-PAGE gel stained with Coomassie blue and in the western blot analysis using AtHD1-C as primary antibody. (C) histone deacetylase assays of purified recombinant AtHD1. rAtHD1 was incubated with synthetic histone H3 or H4 peptides labeled with 3H-acetyl groups on specific lysine residues. The amount of 3H-acetate released due to histone deacetylase activity was measured by scintillation counting. Relative ratios were calculated based on the counts from the control (water + SB = 1). The error bars represent the average values ± S.D. of four duplicates of the assays using independent samples. SB: sodium butyrate.
To determine histone deacetylase activity in vitro, the recombinant AtHD1 was incubated with synthetic histone H3 or H4 peptides labeled with 3H-acetyl groupsonspecific lysine residues. The radiolabeled histone peptides were immobilized on agarose beads so that the histone deacetylase activity of the recombinant AtHD1, if any, would be correlated with the amount of 3H-acetyl groups released from the histone peptides to the supernatant. The HeLa nuclear extract that contains a blend of HDACs was served as the positive control, while sodium butyrate that is a specific specific inhibitor of HDACs was added to the triplicates of each HDAC sample. The amount of 3H-acetate released to the supernatant was measured by scintillation counting as shown in Figure 4C. Compared with the negative control (water) or specificity control (sodium butyrate), the addition of purified rAtHD1 (to a final concentration of 5 mg/ml) led to an increased amount (1.5-fold) of 3H-acetate release, indicative of histone deacetylase activity. The HeLa nuclear extract gave an average of 3.5-fold increase in radioactive count per minute (cpm). The addition of sodium butyrate, a specific inhibitor of histone deacetylases, prevented the reactions containing HeLa nuclear extracts and rAtHD1. Recombinant HDACs (AtHD1, AtHDA6 and AtHDA7) produced in insect cells did not show measurable histone deacetylase activity (data not shown).
Discussion
AtHD1 and other RPD3-like HDACs are differentially expressed in Arabidopsis
Consistent with the data on AtHD1 expression analyzed by RNA gel blot experiments [32] (Plant Chromatin Database, http://chromdb.org) and RT-PCR [27], AtHD1 is expressed in all tissues examined (data not shown). The constitutive expression pattern of AtHD1 suggests that it may have a role in global regulation of plant gene expression [26]. AtHD1 has been shown to regulate gene expression in house-keeping functions such as pathogen response [27] and in floral development such as the expression of SUPERMAN [25] in Arabidopsis. In contrast, the expression of HDA7, which had not been detected in previous studies, was found to express at very low levels in the flower tissues. This flower-specific expression pattern suggests that HDA7 may play a role in flowering. Interestingly, recent studies have shown that histone deacetylation and histone H3 dimethylation at lysines 9 and 27 are involved in FLC (Flowering Locus C) repression, whereas histone acetylation and histone H3 trimethylation at lysine 4 are associated with active FLC expression [33-35]. Therefore, it would be interesting to determine the effects of HDA7 on the expression of flower specific genes.
AtHD1 is localized in the nucleus and excluded from the nucleolus
AtHD1 is supposed to be a nuclear protein that modifies its substrate histones in the chromatin. However, some HDACs can be shuttled between the nucleus and cytoplasm as a way to regulate gene expression. For instance, the mammalian class II HDACs, HDAC4, 5, 7 and 9, are associated with 14-3-3 proteins upon phosphorylation of conserved serine residues on their N-terminal regions, resulting in sequestration of the HDACs to the cytoplasm [36-40]. In contrast, the class I HDACs, including HDAC1, 2 and 8, are predominantly localized in the nucleus and seem not to be regulated by subcellular localization [41]. Exceptionally, class I HDAC3 is present in both the cytoplasm and the nucleus. It is translocated from the nucleus to the cytoplasm in response to interleukin-1b signaling, resulting in derepression of a specific subset of NF-κB regulated genes [42]. In addition, the study of the expression profile and cellular localization of three RPD3-type HDACs in maize, termed ZmRpd3/101, ZmRpd3/102 and ZmRpd3/108, showed that they are present in both cytoplasm and nuclei during different stages of kernel, shoot and anther development [43]. This suggests that the three maize class I HDACs may be regulated by nuclear cytoplasmic shuttling and involved extensively in regulation of various physiological and developmental processes [43].
We investigated subcellular localization of AtHD1 extensively by examining the expression of AtHD1-GFP fusion protein in various intact living tissues of Arabidopsis with high resolution. The results of transient and stable expression of AtHD1-GFP fusion protein indicated the localization of AtHD1 in the cell nuclei of onion epidermal cells and all Arabidopsis tissues examined, including root, leaf, flower and seed. Further examination of the AtHD1-GFP localization patterns indicates that the protein is likely excluded from the nucleolus, which suggests a role of AtHD1 in gene regulation that is different from ZmHD2, AtHD2 and AtHDA6 [8, 20, 23]. The GFP protein used in this experiment is an engineered one called S65T GFP that contains a mutation in the chromophore with the serine 65 being replaced by threonine, resulting in enhanced brightness, faster chromophore formation, and slower photobelaching [44]. Control experiments with the expression of non-fusion GFP protein were not included in this study because the S65T and other types of GFP proteins without a targeting sequence had been shown to accumulate and distribute evenly in Arabidopsis and onion cells due to their small sizes [31, 45, 46]. In fact, the size exclusion limit of the nuclear pores has been estimated to be 40 to 60 kDa [47, 48]. Therefore, GFP protein having a molecular weight of 27 kDa can diffuse across the nuclear envelope, whereas AtHD1-GFP fusion protein that is 83 kDa in size should remain in the cytoplasm if AtHD1 lacks a nuclear localization signal.
The role of histone deacetylation in mitosis
Histone deacetylation was shown to correlate with chromatin condensation and subsequent chromatid segregation during mitosis [49-51]. It has been demonstrated that the histone acetylation levels in several mammalian cell lines started to decrease as the cells enter prophase, then dramatically reduced as the cells progressed into metaphase through telophase, and restored to normal levels at the late telophase/early interphase boundary [49]. Moreover, a temporal inhibition of HDACs by trichostatin A (TSA) in human fibroblasts entering mitosis resulted in defective chromatin condensation and impaired chromatid segregation [50]. A similar study in plant also showed that TSA treatment on cultured tobacco cells led to abnormal anaphases and impaired the progression through mitosis [51]. When we examined the subcellular localization of AtHD1-GFP fusion protein, we captured an image of a leaf mesophyll cell entering mitosis. The cell was at early prophase as evident by the presence of condensing chromatin and the nucleolus was still intact. The GFP signals revealed that AtHD1 aggregated as dots and speckles, suggesting that AtHD1 may be associated with the condensing chromatin during mitosis. We also observed that the GFP signals and DAPI staining did not overlap perfectly, suggesting that AtHD1 might be targeted to specific chromosomal regions.
Recombinant AtHD1 demonstrated histone deacetylase activity in vitro
The histone deacetylase activity of AtHD1 is implied from studies in which AtHD1 was either knocked out or overexpressed. Down-regulation of AtHD1 led to the accumulation of hyperacetylated histones in transgenic Arabidopsis [25], and overexpression of AtHD1 decreased levels of tetraacetylated histone [27]. Here, we demonstrated that the recombinant AtHD1 produced in bacteria showed a moderate yet specific histone deacetylase activity in vitro, providing experimental evidence for enzymatic activity of a plant histone deacetylase. AtHD1 showed enzyme activity in three replicated experiments (using different protein extractions), whereas the recombinant AtHDA6, 9, and 17 did not show enzymatic activity (data not shown), suggesting that the low activity of AtHD1 is not due to contamination of bacterial proteins or artifacts.
The sub-optimal activity of the recombinant AtHD1 may suggest that it requires post-translational modifications or specific chaperons for proper folding, both of which are absent in the bacterial expression system. Phosphorylation is a major post-translation modification in HDACs. In the maize embryos, phosphorylation of HD1A causes a change insubstratespecificity [52]. Other HDACs, such as the maize HD2 [8], human HDAC1, 2, 4 and 5 [53-55] are subjected to phosphorylation, which can modulate their activities. Another post-translational modification that has been shown to regulate HDAC activity and function is called sumoylation, which involves the conjugation of small ubiquitin-related modifier [41, 56-58]. However, sumoylation has been identified only in mammalian HDACs. Some HDACs are regulated by proteolytic processing. In maize, HDA1 is synthesized as an inactive precursor protein that is converted to the enzymatically active form by proteolytic removal of the C-terminal part of the protein [59]. In an attempt to solve the problem caused by the lack of post-translational modifications, AtHD1 together with other RPD3-type HDACs in Arabidopsis including HDA6, 7 and 9, were produced by baculoviral expression system and then tested by HDAC assay. However, all recombinant HDACs tested did not show any HDAC activity (data not shown).
A second possible reason for the low activity of rAtHD1 is lack of co-factors or requirement of complex formation with other proteins. Notably, many purified recombinant HDACs are enzymatically inactive [41], with only the exception of yeast HOS3 and mammalian HDAC8 [60-62]. It will be interesting to further investigate AtHD1 activity by isolating interacting proteins.
Acknowledgments
We thank Mary Bryk and Timothy Hall for critical suggestions to improve the manuscript, David Stelly and Keerti Rathore for assistance in GFP localization studies in onion cells, and Stanislav Vitha in the Microscopy and Imaging Center at Texas A&M University for technical support for epifluorescence microscopic image analysis in the transgenic plants. The work is supported by grants from the National Institutes of Health (GM067015) and the National Science Foundation Plant Genome ResearchProgram (DBI0077774) to Z J C.
References
- 1.Brownell JE, Allis CD. Special HATs for special occasions: linking histone acetylation to chromatin assembly and gene activation. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 2.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 3.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 4.Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–511. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- 5.Pandey R, Muller A, Napoli CA, et al. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rundlett SE, Carmen AA, Kobayashi R, et al. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci U S A. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loidl P. A plant dialect of the histone language. Trends Plant Sci. 2004;9:84–90. doi: 10.1016/j.tplants.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 8.Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- 9.Alland L, Muhle R, Hou H, Jr, et al. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- 10.Hassig CA, Fleischer TC, Billin AN, Schreiber SL, Ayer DE. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell. 1997;89:341–347. doi: 10.1016/s0092-8674(00)80214-7. [DOI] [PubMed] [Google Scholar]
- 11.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deacetylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 12.Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenzie EA, Kent NA, Dowell SJ, et al. The centromere and promoter factor, 1, CPF1, of Saccharomyces cerevisiae modulates gene activity through a family of factors including SPT21, RPD1 (SIN3), RPD3 and CCR4. Mol Gen Genet. 1993;240:374–386. doi: 10.1007/BF00280389. [DOI] [PubMed] [Google Scholar]
- 14.Stillman DJ, Dorland S, Yu Y. Epistasis analysis of suppressor mutations that allow HO expression in the absence of the yeast SW15 transcriptional activator. Genetics. 1994;136:781–788. doi: 10.1093/genetics/136.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jackson JC, Lopes JM. The yeast UME6 gene is required for both negative and positive transcriptional regulation of phospholipid biosynthetic gene expression. Nucleic Acids Res. 1996;24:1322–1329. doi: 10.1093/nar/24.7.1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Nadal E, Zapater M, Alepuz PM, et al. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004;427:370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- 17.Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 2001;29:1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HJ, Hyun Y, Park JY, et al. A genetic link between cold responses and flowering time through FVE in Arabidopsis thaliana. Nat Genet. 2004;36:167–171. doi: 10.1038/ng1298. [DOI] [PubMed] [Google Scholar]
- 19.Rossi V, Hartings H, Motto M. Identification and characterisation of an RPD3 homologue from maize (Zea mays L.) that is able to complement an rpd3 null mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1998;258:288–296. doi: 10.1007/s004380050733. [DOI] [PubMed] [Google Scholar]
- 20.Murfett J, Wang XJ, Hagen G, Guilfoyle TJ. Identification of Arabidopsis histone deacetylase hda6 mutants that affect transgene expression. Plant Cell. 2001;13:1047–1061. doi: 10.1105/tpc.13.5.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aufsatz W, Mette MF, Van Der Winden J, Matzke M, Matzke AJ. HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;2002:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:E67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Probst AV, Fagard M, Proux F, et al. Arabidopsis histone deacetylase HDA6 is required for maintenance of transcriptional gene silencing and determines nuclear organization of rDNA repeats. Plant Cell. 2004;16:1021–1034. doi: 10.1105/tpc.018754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tian L, Wang J, Fong MP, et al. Genetic control of developmental changes induced by disruption of Arabidopsis histone deacetylase 1 (AtHD1) expression. Genetics. 2003;165:399–409. doi: 10.1093/genetics/165.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tian L, Chen ZJ. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci U S A. 2001;98:200–205. doi: 10.1073/pnas.011347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian L, Fong MP, Wang JJ, et al. Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics. 2005;169:337–345. doi: 10.1534/genetics.104.033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 Is Involved in Jasmonic Acid and Ethylene Signaling of Pathogen Response in Arabidopsis. Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clough SJ, Bent AF. Floral dip: a simlified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 29.Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- 30.Bent AF, Clough SJ. Agrobacterium germ-line transformation: Transfromation of Arabidopsis without tissue culture. In: Gelvin SB, Schilperoort RA, editors. Plant Molecular Biology Manual. Kluwer Academic Pub; Dordrecht, The Netherlands: 1998. pp. 1–14. [Google Scholar]
- 31.Lawrence RJ, Earley K, Pontes O, et al. A concerted DNA methylation/histone methylation switch regulates rRNA gene dosage control and nucleolar dominance. Mol Cell. 2004;13:599–609. doi: 10.1016/s1097-2765(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 32.Wu K, Malik K, Tian L, Brown D, Miki B. Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol Biol. 2000;44:167–176. doi: 10.1023/a:1006498413543. [DOI] [PubMed] [Google Scholar]
- 33.He Y, Michaels SD, Amasino RM. Regulation of flowering time by histone acetylation in Arabidopsis. Science. 2003;302:1751–1754. doi: 10.1126/science.1091109. [DOI] [PubMed] [Google Scholar]
- 34.Ausin I, Alonso-Blanco C, Jarillo JA, Ruiz-Garcia L, Martinez-Zapater JM. Regulation of flowering of time by FVE, a retinoblastoma-associated protein. Nat Genet. 2004;36:162–166. doi: 10.1038/ng1295. [DOI] [PubMed] [Google Scholar]
- 35.He Y, Amasino RM. Role of chromatin modification in flowering-time control. Trends Plant Sci. 2005;10:30–35. doi: 10.1016/j.tplants.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Grozinger CM, Schreiber SL. Regulation of histone deacetylase 4 and 5 and transcriptional activity by 14-3-3-dependent cellular localization. Proc Natl Acad Sci U S A. 2000;97:7835–7840. doi: 10.1073/pnas.140199597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKinsey TA, Zhang CL, Olson EN. Activation of the myocyte enhancer factor-2 transcription factor by calcium/calmodulin-dependent protein kinase-stimulated binding of 14-3-3 to histone deacetylase 5. Proc Natl Acad Sci U S A. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang AH, Kruhlak MJ, Wu J, et al. Regulation of histone deacetylase 4 by binding of 14-3-3 proteins. Mol Cell Biol. 2000;20:6904–6912. doi: 10.1128/mcb.20.18.6904-6912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao HY, Verdel A, Tsai CC, et al. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J Biol Chem. 2001;276:47496–47507. doi: 10.1074/jbc.M107631200. [DOI] [PubMed] [Google Scholar]
- 40.Miska EA, Karlsson C, Langley E, et al. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. Embo J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem. 2004;93:57–67. doi: 10.1002/jcb.20179. [DOI] [PubMed] [Google Scholar]
- 42.Baek SH, Ohgi KA, Rose DW, et al. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-kappaB and beta-amyloid precursor protein. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 43.Varotto S, Locatelli S, Canova S, et al. Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol. 2003;133:606–617. doi: 10.1104/pp.103.025403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiu W, Niwa Y, Zeng W, et al. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 46.Koroleva OA, Tomlinson ML, Leader D, Shaw P, Doonan JH. High-throughput protein localization in Arabidopsis using Agrobacterium-mediated transient expression of GFP-ORF fusions. Plant J. 2005;41:162–174. doi: 10.1111/j.1365-313X.2004.02281.x. [DOI] [PubMed] [Google Scholar]
- 47.Grebenok RJ, Pierson E, Lambert GM, et al. Green-fluorescent protein fusions for efficient characterization of nuclear targeting. Plant J. 1997;11:573–586. doi: 10.1046/j.1365-313x.1997.11030573.x. [DOI] [PubMed] [Google Scholar]
- 48.Haasen D, Kohler C, Neuhaus G, Merkle T. Nuclear export of proteins in plants: AtXPO1 is the export receptor for leucine-rich nuclear export signals in Arabidopsis thaliana. Plant J. 1999;20:695–705. doi: 10.1046/j.1365-313x.1999.00644.x. [DOI] [PubMed] [Google Scholar]
- 49.Kruhlak MJ, Hendzel MJ, Fischle W, et al. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- 50.Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol Biol Cell. 2003;14:3821–3833. doi: 10.1091/mbc.E03-01-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li DW, Yang Q, Chen JT, et al. Dynamic distribution of Ser-10 phosphorylated histone H3 in cytoplasm of MCF-7 and CHO cells during mitosis. Cell Res. 2005;15:120–126. doi: 10.1038/sj.cr.7290276. [DOI] [PubMed] [Google Scholar]
- 52.Brosch G, Georgieva EI, Lopez-Rodas G, Lindner H, Loidl P. Specificity Zea mays histone deacetylase is regulated by phosphorylation. J Biol Chem. 1992;267:20561–20564. [PubMed] [Google Scholar]
- 53.Cai R, Kwon P, Yan-Neale Y, et al. Mammalian histone deacetylase 1 protein is posttranslationally modified by phosphorylation. Biochem Biophys Res Commun. 2001;283:445–453. doi: 10.1006/bbrc.2001.4786. [DOI] [PubMed] [Google Scholar]
- 54.Pflum MK, Tong JK, Lane WS, Schreiber SL. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem. 2001;276:47733–47741. doi: 10.1074/jbc.M105590200. [DOI] [PubMed] [Google Scholar]
- 55.Tsai SC, Seto E. Regulation of histone deacetylase 2 by protein kinase CK2. J Biol Chem. 2002;277:31826–31833. doi: 10.1074/jbc.M204149200. [DOI] [PubMed] [Google Scholar]
- 56.Colombo R, Boggio R, Seiser C, Draetta GF, Chiocca S. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 2002;3:1062–1068. doi: 10.1093/embo-reports/kvf213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.David G, Neptune MA, DePinho RA. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem. 2002;277:23658–663. doi: 10.1074/jbc.M203690200. [DOI] [PubMed] [Google Scholar]
- 58.Kirsh O, Seeler JS, Pichler A, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. Embo J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pipal A, Goralik-Schramel M, Lusser A, et al. Regulation and processing of maize histone deacetylase Hda1 by limited proteolysis. Plant Cell. 2003;15:1904–1917. doi: 10.1105/tpc.013995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carmen AA, Griffin PR, Calaycay JR, et al. Yeast HOS3 forms a novel trichostatin A-insensitive homodimer with intrinsic histone deacetylase activity. Proc Natl Acad Sci U S A. 1999;96:12356–12361. doi: 10.1073/pnas.96.22.12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu E, Chen Z, Fredrickson T, et al. Cloning and characterization of a novel human class I histone deacetylase that functions as a transcription repressor. J Biol Chem. 2000;275:15254–15264. doi: 10.1074/jbc.M908988199. [DOI] [PubMed] [Google Scholar]
- 62.Lee H, Rezai-Zadeh N, Seto E. Negative regulation of histone deacetylase 8 activity by cyclic AMP-dependent protein kinase A. Mol Cell Biol. 2004;24:765–773. doi: 10.1128/MCB.24.2.765-773.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


