Abstract
Anginex, a designed peptide 33mer, is known to function both as an antiangiogenic and bactericidal agent. Solving the NMR solution structure of the peptide is key to understand better its structure-activity relationships and to design more bioactive peptides and peptide mimetics. However, structure elucidation of anginex has been elusive due to subunit exchange-induced resonance broadening. Here, we found that performing NMR structural studies in a micellar environment abolishes exchange broadening and allows the structure of anginex to be determined. Anginex folds in an amphipathic, three-stranded antiparallel β-sheet conformation with functionally key hydrophobic residues lying on one face of the β-sheet and positively charged, mostly lysine residues, lying on the opposite face. Structural comparison is made with a homologous, yet relatively inactive peptide, βpep-28. These results contribute to the design of peptidomimetics of anginex for therapeutic use against angiogenically-related diseases like cancer, as well as infectious diseases.
Keywords: drug design, anti-angiogenic, anti-bacterial, conformation
INTRODUCTION
Angiogenesis, the process by which new vessels form from pre-existing vessels [1], is essential for various bodily functions, like reproduction, embryogenesis, and tissue repair. It is also fundamental to pathological diseases, such as rheumatoid arthritis, retinopathy, and tumor growth [2]. Anti-angiogenic therapy is currently considered to be a promising approach to therapeutic intervention, as shown by in vitro and in vivo anti-tumor research studies over the past several years [3-6]. Curiously, there appears to be a general correlation between antiangiogenic and bactericidal proteins, in that an antiangiogenic protein usually demonstrates some bactericidal activity [7]. Although an evolutionary link is likely, the actual reason for this remains unclear. There is certainly a structural link in that these peptides contain primarily β-sheet structure. It is also known that peptides in this class kill bacteria by integrating into bacterial membranes and forming leaky channels indiscriminately, hence their general name of membrane disintegrating peptides.
Most anti-angiogenic proteins have been identified by isolating endogenous molecules which inhibit endothelial cell (EC) growth, e.g. platelet factor-4 (PF4) [8], thrombospondin-1 [9], angiostatin [10], endostatin [11], and bactericidal-permeability increasing (BPI) protein [12]. Previously, we reported the design of a series of β-sheet-forming peptide 33mers (βpep peptides) [13], all of which demonstrate bactericidal activity to varying degrees [14] and a few of which elicite anti-angiogenic activity [15]. The most anti-angiogenically potent of these peptides is βpep-25 (anginex), which effectively inhibits tumor angiogenesis and tumor growth [15-18]. The molecular target of anginex was recently identified as galectin-1 (gal-1) [19], a protein (highly upregulated by tumor endothelium) that is crucial to endothelial cell adhesion and migration, and therefore to tumor angiogenesis. Anginex (βpep-25), a gal-1 antagonist, binds gal-1 strongly (Kd = 90 nM) and specifically (1:1). In addition, anginex also interacts weakly (Kd = 20 μM) and apparently non-specifically (about 50:1) with plasma fibronectin [20], an interaction that likely promotes the peptide’s transport through the cardiovascular system to the tumor vasculature where it then binds gal-1.
Figure 1 displays the amino acid sequences of anginex (βpep-25) and a homologous, yet relatively anti-angiogenically inactive peptide, βpep-28. A survey of amino acid sequences from numerous anti-angiogenic proteins reveals that they are compositionally similar, containing numerous hydrophobic and cationic residues [7,16]. These structural and compositional characteristics, which appear to be functionally important, are embodied in anginex [15]. Circular dichroism (CD) data on anginex have indicated the presence of significant populations of β-sheet [15, 21]. Although the NMR structure of another βpep peptide, βpep-4, has been solved [22], elucidation of the anginex structure by NMR has been difficult, primarily because all βpep peptides self-associate, usually as dimers and tetramers [13,14], and, depending on the subunit-exchange rate, resonance broadening occurs [15]. For anginex, unfortunately, the subunit exchange rate falls within the time regime where broadening is greatest, making NMR spectral sensitivity and resolution extremely poor. Recently, we found that this resonance broadening effect can be overcome by studying anginex in the presence of detergent micelles, which tend to shift the subunit exchange rate into a regime where broadening is no longer problematic and resonances are well defined. Use of such a model system was deemed relevant because biologically, anginex interacts with bacterial membranes to function as a bactericidal agent [14,23], as well as with eukaryotic membranes as the peptide interacts with its adhesion/migration-mediating receptor [15,19].
Figure 1.
Amino acid sequence of anginex and βpep-28. The amino acid sequences of anginex and homologous peptide βpep-28 are shown.
The present study was aimed at determining the NMR solution structures of anginex and homologous, yet antiangiogenically-inactive, control peptide βpep-28 [15] in a DPC micelle system. Here, we report that both anginex and βpep-28 fold as amphipathic, three-stranded antiparallel β-sheets. Comparison of their structures provides insight into which structural features and positions of key amino acid residues help to impart biological activity to anginex. Knowledge of the folded structure of anginex is considered key to understand structure-activity relationships and to design more bioactive peptides and peptide mimetics.
EXPERIMENTAL PROCEDURES
Peptide synthesis
βpep peptide 33mers (anginex and βpep-28) were synthesized and HPLC purified as previously reported [13,14]. Analytical HPLC, mass spectrometery, and N-terminal sequencing confirmed greater than 95% purity of the peptides.
NMR Measurements
Freeze-dried anginex or βpep-28 was dissolved in a mixture of H2O/DMSO (1:1, v/v) in the presence or absence of 50 mM perdeuterated (D38) DPC micelles (Cambridge Isotope Labs, Inc., Andover, MA). For most of the NMR experiments, peptide concentration was approximately 1 mM. The pH was adjusted to pH 5.5 by adding μL quantities of NaOH or HCl to the peptide sample. NMR spectra were acquired at 40 °C on a Varian UNITY Plus-600 NMR spectrometer, using a homonuclear NMR approach previously described [22]. Data were processed offline using the program NMRPipe [24] on an SGI workstation and were analyzed using using the program Sparky (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco). Resonance assignments were made for both peptides in the presence or absence of DPC micelles, as stated above. Chemical shift differences (Δδ) between resonances of the same group were calculated as the chemical shift of the group in DPC micelles minus the chemical shift of that group in the absence of DPC micelles.
Structural Modeling
Analysis of nuclear Overhauser effect (NOE) growth curves indicated that backbone-to-backbone interproton NOEs were maximum at about 200 ms. Interproton distance constraints were derived from NOEs assigned in 1H NOE spectra acquired with mixing times of 200 ms. NOEs were classified as strong, medium, weak, or very weak corresponding to upper bound distance constraints of 2.8 Å, 3.3 Å, 4.0 Å, and 5.0 Å, respectively. The lower bound restraint between non-bonded protons was set at 1.8 Å. Pseudo-atom corrections were added to the upper bound distance constraints where appropriate, and a 0.5 Å correction was added to the upper bound for NOEs involving methyl protons. Hydrogen bond constraints were identified from the pattern of sequential and interstrand NOEs involving NH and CαH protons together with evidence of slow amide proton-solvent exchange. Each hydrogen bond identified was defined using two distance constraints: rNH-O = 1.8−2.5 Å and rN-O = 2.5−3.3 Å. The program X-PLOR [25] was used to calculate structures for anginex and βpep-28 using derived internuclear distance constraints as previously described [26]. Calculated structures were superimposed using the programs MolMol [27] and Insight II (Accelrys, Inc., San Diego, CA) and were analyzed using X-PLOR analysis routines.
RESULTS & DISCUSSION
In a solution of only H2O/DMSO (1:1), backbone resonances from both anginex and βpep-28 exhibit chemical shifts characteristic of random coil conformation [28]. In the presence of DPC micelles, however, both peptides display NMR spectra showing well dispersed, downfield shifted αH and NH resonances characteristic of β-sheet conformation [28]. To exemplify this, Figure 2 shows the αH-NH region from a TOCSY spectrum of anginex (Figure 2A), along with that of homologous peptide βpep-28 (Figure 2C). Sequential assignments were done by using standard two-dimensional 1H NMR methods [29], and some cross-peak assignments are indicated in the figure.
Figure 2.
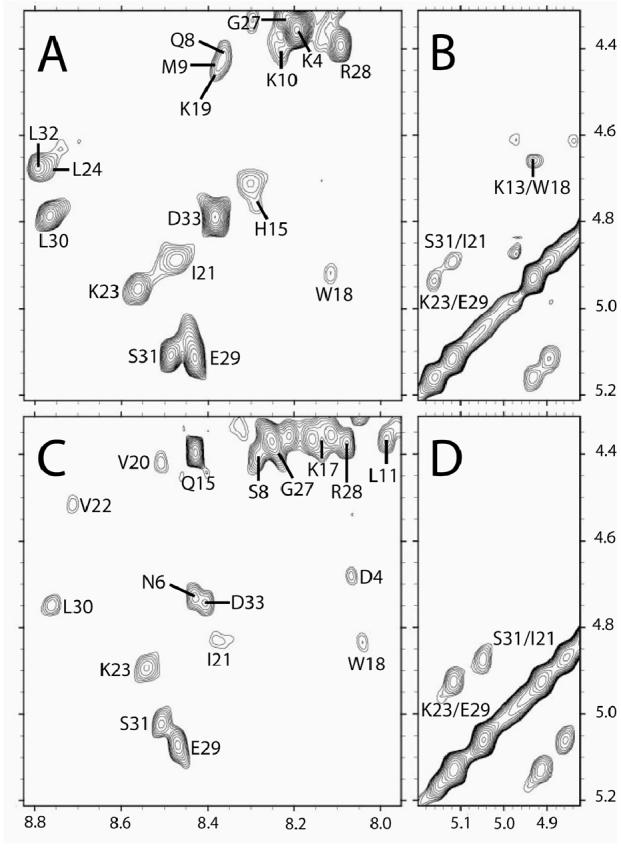
TOCSY and NOESY spectra for anginex and βpep-28. 600 MHz 1H 60 ms TOCSY and 200 ms NOESY spectra are shown for anginex (A, B) and βpep-28 (C, D). Peptide concentration was 1.0 mM in H2O/DMSO (1:1) and 50 mM DPC, pH 5.5 and 40 °C. Spectra were accumulated with 4 k data points over 9000 Hz sweep width and were processed with 1 Hz line broadening. Only spectral regions downfield from the HDO resonance are shown and some key NOEs are identified. Several cross-peaks have been labeled. Some cross-strand αH-αH NOEs used to indicate β-strand alignment are also indicated.
Exemplary evidence for the presence of anti-parallel, β-sheet conformation in anginex and in βpep-28 is provided by NOESY data showing αH-αH cross-strand NOEs (Figures 2B and 2D, respectively). Other cross-strand backbone-to-backbone NOEs observed from anginex are indicated by arrows in Figure 3A, along with the alignment of the three β-strands. A summary of other NOEs observed from anginex is given in Figure 3B. Similar results were observed for βpep-28 (data not shown), which shows the same alignment of the three β-strands.
Figure 3.
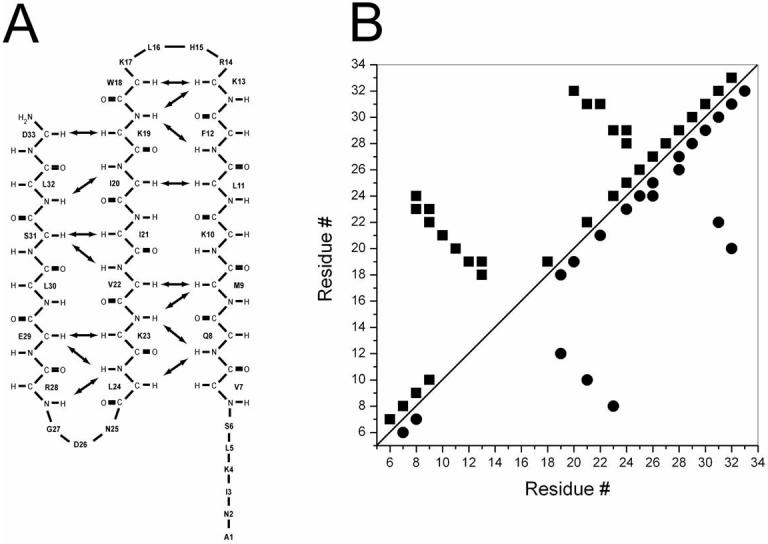
Summary of NOEs and β-strand alignments in anginex. The alignment of the three β-strands of anginex is shown, along with key cross-strand backbone-to-backbone NOEs indicated by arrows (A). A summary of observed NOEs for anginex is also shown (B). In panel B, backbone to backbone NOEs are shown with squares in the upper triangle, and backbone to side chain and side chain to side chain NOEs are shown with circles in the lower triangle. Correlations indicated are for all NOEs for any given residue.
It is interesting to note that NOEs between β-strands 2 and 3 are generally stronger than those between β-strands 1 and 2. Note, however, that the K13-W18 cross-strand αH-αH NOE displays essentially the same magnitude as αH-αH NOES between strands 2 and 3. Cross-strand NOEs tend to be weaker for more N-terminal residues. Although the reason for this is unclear, it is probably due to motional dynamics and/or greater N-terminal interactions with the DPC micelles. This trend does appear to parallel that observed with αH and NH chemical shift differences (Δδ) illustrated in Figures 4A (anginex) and 4B (βpep-28). Δδ values were calculated as the chemical shift in the presence of DPC micelles minus the chemical shift in their absence. Wishart et al. [30] reported that downfield chemical shift differences relative to random coil positions indicate β-sheet formation, whereas upfield shift differences indicate helix formation. Particularly with peptides where equilibrium among conformational states is likely, one could say that the greater the shift difference is, the more stable that particular conformation is. For β-strands 2 and 3 in both anginex and βpep-28, αH and NH chemical shifts in DPC micelles are more downfield shifted (i.e., Δδ values in Figure 4 are more positive), especially for residues 20 to 24 and 29 to 32 (Figures 4A and 4B). For β-strand 1, on the other hand, αH and NH chemical shift differences tend to be more negative (upfield shifted), and even though as mentioned above β-sheet-type NOEs from residues in β-strand 1 are generally somewhat weaker than those from β-strands 2 and 3, there is no evidence for NOEs characteristic of helix formation in β-strand 1. Moreover, interaction with micelles could also contribute to chemical shifts of resonances belonging to residues in β-strand 1. Resonances belonging to residues in the first turn connecting strands 1 and 2 (residues 14−17) were broad and could not be uniquely identified, whereas those from the second turn (residues 25−27) are mostly negative (upfield shifted), consistent with a well-formed, tight turn.
Figure 4.
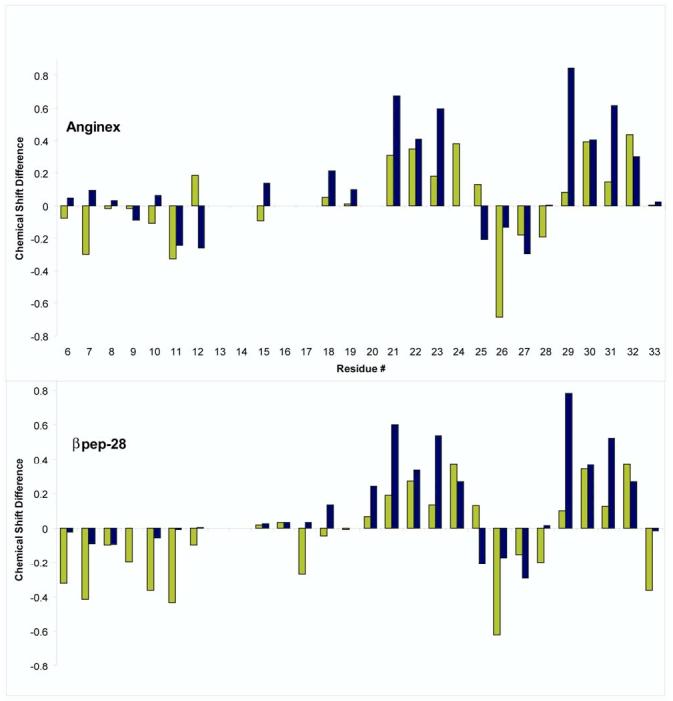
Backbone chemical shift differences for anginex and βpep-28. Chemical shift differences are defined here as the difference in chemical shifts of resonances from the peptides in the presence of DPC micelles (50 mM DPC in 1:1 H2O/DMSO) from chemical shifts of resonances from the peptides in 1:1 H2O/DMSO alone. Chemical shifts are taken from TOCSY spectra collected at 600 MHz and 40 °C for resonances that could be unambiguously identified. Backbone NH and αH chemical shift differences (ΔδHN and ΔδHA) for anginex and βpep-28 are shown in panels A and B, respectively. ΔδHN and ΔδHA values are given in bar graph format in gray and black shading, respectively.
Regardless of interaction of the peptides with these micelles, relatively complete sets of NOEs for only one structural type for both anginex and βpep-28 were observed. Because of this, conformational modeling was performed using these NOESY data. For anginex, there were a total of 236 NOE distance constraints, including 161 intraresidue, 14 sequential, 40 medium-range ( | i - j | < 5), and 21 long-range ( | i - j| > 5) constraints. For βpep-28, there were a total of 294 NOE distance constraints, including 201 intraresidue, 21 sequential, 45 medium-range ( | i - j | < 5), and 27 long-range ( | i - j| > 5) constraints. In addition, a total of 12 hydrogen bonds could be identified for either peptide via inspection of initial structures and from the presence of long-lived backbone NHs, giving rise to 24 hydrogen bond distance constraints. Therefore, the total number of experimentally derived constraints was 248 for anginex (average of 8 constraints per residue) and 306 for βpep-28 (average of 10 constraints per residue).
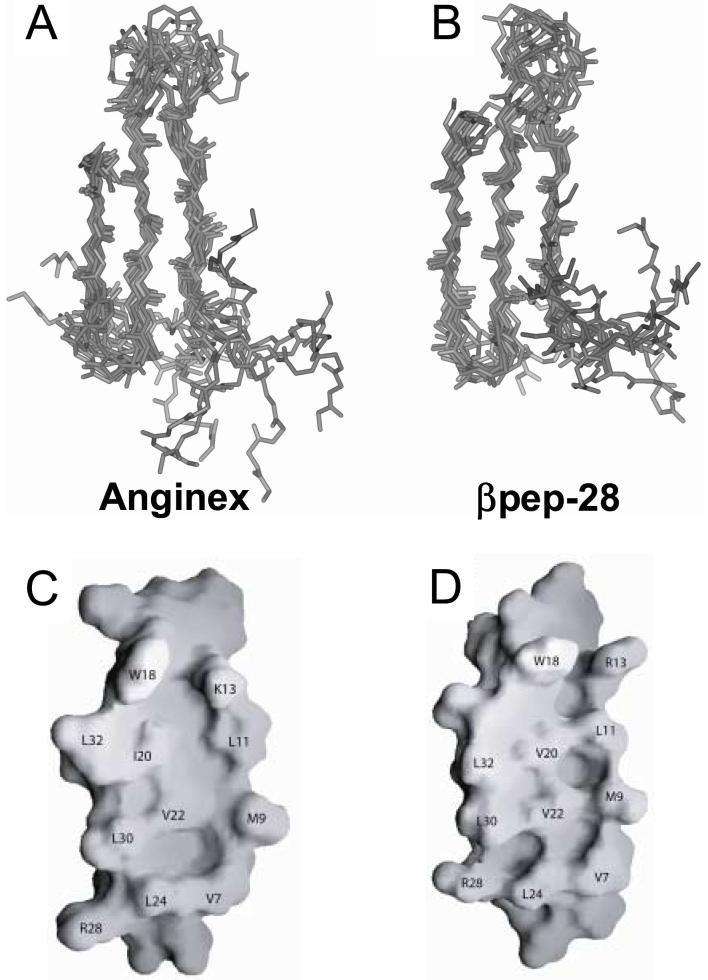
Initially, 100 structures for each peptide were calculated as described in the Methods Section. The best fit superpositions of backbone Cα atoms for the final 13 and 11 structures are shown for anginex and for βpep-28, respectively, in Figures 5A and 5B. These structures have no NOE violations greater 0.5 Å. Structural statistics are given in Table 1, and show that the structures satisfy experimental constraints quite well. In addition, ϕ and ψ angular order parameters are all > 0.8. Taken together, the above data indicate that structures used to represent the solution conformations of anginex and βpep-28 are well converged. For anginex, the RMSD for backbone and heavy atoms of β-sheet residues 8−13, 18−23 and 29−32 (excludes termini and loop residues) is 0.52 Å and 1.34 Å, respectively, whereas for βpep-28, these RMSD values are 0.73 Å and 1.79 Å, respectively. In either peptide, the N-terminus is generally more poorly defined. In fact, residues 2 − 5 in anginex, and residues 1, 2 and 5 in βpep-28, could not be uniquely identified, thus accounting for the higher degree of randomness in the N-termini.
Figure 5.
Structures and hydrophobic surfaces of anginex and βpep-28. The best fit superpositions of backbone Cα atoms for the final 13 and 11 NMR-derived structures are shown for anginex (A) and for βpep-28 (B), respectively. From these NMR-derived structures, the hydrophobic surfaces of anginex (C) and βpep-28 (D) are shown for the average structures, and some residues are labeled. In panels C and D, the flexible N-terminus is omitted from the surface shown.
Table 1.
Structural statistics for the calculated structures of anginex and βpep-28 from NMR data.
| RMS Deviations from experimental distance restraints (Å)a | ||
| NOE (260, 318) | anginex | βpep-28 |
| H-bond (24) | 0.058 ± 0.006 | 0.069 ± 0.005 |
| Deviations from idealized geometry | ||
| Bonds (Å) | 0.003 | 0.004 |
| Angles (°) | 0.57 ± 0.03 | 0.58 ± 0.02 |
| Impropers (°) | 0.45 ± 0.03 | 0.42 ± 0.03 |
| Energies (kcal.mol−1) | ||
| ENOEb | 41 ± 8 | 64 ± 9 |
| EBOND | 7 ± 2 | 9 ± 1 |
| EANGLE | 54 ± 6 | 54 ± 5 |
| EVDW | 14 ± 4 | 12 ± 4 |
| EIMPROPER | 9 ± 1 | 7 ± 1 |
| ETOTAL | 125 ± 15 | 147 ± 16 |
None of the final 13 structures of anginex or final 11 structures of βpep-28 exhibited distance restraint violations greater than 0.5 Å or dihedral angle violations greater than 5°. RMSD values represent the mean and standard deviations for the structures.
The final value of the NOE (ENOE) was calculated with a force constant of 50 kcal.mol−1.Å−2.
Although anginex and βpep-28 have essentially the same structure, there are some minor differences. Structural comparison shows an average RMSD difference of 1.27 Å for backbone atoms over the three β-strands (8−13, 18−23, 29−32). In βpep-28, the first turn connecting strands 1 and 2 appears to be less tight than that in anginex, whereas the second turn connecting strands 2 and 3 in βpep-28 is slightly more twisted. Given that these are relatively flexible peptides and unlike more stable folds in proteins, it is likely that these minor differences are merely the result of computational artifacts and do not contribute to understanding differences in their bioactivities.
Both peptides also fold similarly to homologous peptide, βpep-4 [22]. However, βpep-4 at 1 mM in water also self-associates into dimers and tetramers via interaction of its β-sheet and hydrophobic face, respectively [13]. Although at 1 mM in water both anginex and βpep-28 also self-associate as monomer, dimers, and tetramers [14,15], they apparently do not aggregate at this peptide concentration under solution conditions used in the present study. NOEs that clearly demonstrated self-association of βpep-4 [16] or βpep-28 [15] in water, are not observed for either anginex or βpep-28 in 1:1 DMSO/H2O solution or in the presence of DPC micelles. Moreover, chemical shifts of resonances from both peptides are essentially the same at the lower peptide concentration of 0.2 mM. Disolution of the peptides in DMSO/H20 and/or by interaction with DPC micelles apparently attenuates βpep peptide self-assembly.
Previously, we demonstrated that residues on the hydrophobic face (V7, L11, I20, V22 and L24) of amphipathic anginex appear to be more functionally important to peptide’s anti-angiogenic activity [31]. Changing residues on the hydrophilic surface is better tolerated, although net positive charge character is also required for activity. Overall, this suggests that the hydrophobic surface of anginex interacts directly with its molecular receptor, galectin-1 [19]. Using our NMR-derived structures, Figure 6 illustrates the surface of the hydrophobic faces of anginex and βpep-28. Residues comprising these surfaces are labeled. Note that because we are dealing with NMR-derived structures of small peptides, side chain positions can vary significantly within each structure due to internal flexibility, and caution should be exercised when comparing these surfaces too exactly. However, it is interesting that only two residues differ between these surfaces (K13 and I20 in anginex, and R13 and V20 in βpep-28). Because βpep-28 is relatively inactive as an antiangiogenic agent compared to anginex and their structures are essentially the same, the activity difference is likely related to these residue differences. We already know that the six C-terminal residues of anginex are dispensible [31].
These structure-activity relationships may be useful to help design a more effective anti-angiogenic agent. For example, primary amines (lysines) vis a vis guanidinium groups (arginines) and isobutyl groups (isoleucine) vis a vis isopropyl groups (valine) may be more desirable to promote anginex-like activity. Based on the amphipathic and cationic character of anti-angiogenic proteins in general, we recently designed a series of non-peptide, calixarene-based protein surface topomimetics [32]. Two of them, 0118 and 1097, display potent antiangiogenic activities. Although functionally 0118 behaves the same as anginex, suggesting the same mechanism of action as anginex (possibly through gal-1), 1097 functions clearly differently. The anti-angiogenic activity of these novel compounds, at least 0118, may be optimized by taking into consideration the chemical character of and the spatial relationship between K13 and I20 in our structure of anginex.
CONCLUSIONS
This NMR study demonstrates that the antiangiogenic and antibacterial peptide anginex forms a three-stranded anti-parallel β-sheet conformation in solution with DPC micelles. Anginex interacts with these micelles and may do so in a similar way when interacting with bacterial and eukaryotic membranes. Using this information, we are currently designing and optimizing non-peptidic molecules to mimic the structure and activity of anginex.
ACKNOWLEDGEMENTS
This work was supported by support from the National Institutes of Health (NIH CA-96090 and AI-057153). NMR instrumentation was provided with funds from the NSF (BIR-961477) and the University of Minnesota Medical School. Peptides were synthesized at the Microchemical Facility, University of Minnesota.
Glossary
Abbreviations
- NMR
nuclear magnetic resonance spectroscopy
- NOE
nuclear Overhauser effect
- NOESY
two-dimensional NOE spectroscopy
- FID
free induction decay
- HPLC
high performance liquid chromatography
- DPC
dodecylphosphocoline
- DMSO
dimethyl sulfoxide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Folkman J. Angiogenesis. Annu Rev Med. 2006;57:1–18. doi: 10.1146/annurev.med.57.121304.131306. [DOI] [PubMed] [Google Scholar]
- 2.Carmeliet P. Angiogenesis in life, disease, and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- 3.Folkman J. Endogenous angiogenesis inhibitors. APMIS. 2004;112:496–507. doi: 10.1111/j.1600-0463.2004.apm11207-0809.x. [DOI] [PubMed] [Google Scholar]
- 4.Folkman J. Angiogenesis inhibitors: a new class of drugs. Cancer Biol. Ther. 2003;2:127–133. [PubMed] [Google Scholar]
- 5.Folkman J. Antiangiogenesis in cancer therapy – endostatin and its mechanism of action. Exp Cell Res. 2006;312:594–607. doi: 10.1016/j.yexcr.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 6.Verhoef C, de Wilt JH, Verheul HM. Angiogenesis inhibitors: perspectives for medical, surgical and radiation oncology. Curr Pharm Des. 2006;12:2623–2630. doi: 10.2174/138161206777698756. [DOI] [PubMed] [Google Scholar]
- 7.Dings RP, Nesmelova I, Griffioen AW, Mayo KH. Discovery and development of anti-angiogenic peptides: A structural link. Angiogenesis. 2003;6:83–91. doi: 10.1023/B:AGEN.0000011730.94233.06. [DOI] [PubMed] [Google Scholar]
- 8.Gupta SK, Singh JP. Inhibition of endothelial cell proliferation by platelet factor-4 involves a unique action on S phase progression. J Cell Biol. 1994;127:1121–1127. doi: 10.1083/jcb.127.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rastinejad F, Polverini PJ, Bouck NP. Regulation of the activity of a new inhibitor of angiogenesis by a cancer suppressor gene. Cell. 1989;56:345–355. doi: 10.1016/0092-8674(89)90238-9. [DOI] [PubMed] [Google Scholar]
- 10.O'Reilly MS, Holmgren L, Shing Y, Chen C, Rosenthal RA, Moses M, Lane WS, Cao Y, Sage EH, Folkman J. Angiostatin: a novel angiogenesis inhibitor that mediates the suppression of metastases by a Lewis lung carcinoma. Cell. 1994;79:315–328. doi: 10.1016/0092-8674(94)90200-3. [DOI] [PubMed] [Google Scholar]
- 11.O'Reilly MS, Boehm T, Shing Y, Fukai N, Vasios G, Lane WS, Flynn E, Birkhead JR, Olsen BR, Folkman J. Endostatin: an endogenous inhibitor of angiogenesis and tumor growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 12.van der Schaft DW, Toebes EA, Haseman JR, Mayo KH, Griffioen AW. Bactericidal/permeability-increasing protein (BPI) inhibits angiogenesis via induction of apoptosis in vascular endothelial cells. Blood. 2000;96:176–181. [PubMed] [Google Scholar]
- 13.Mayo KH, Ilyina E, Park H. A recipe for designing water-soluble, β-sheet-forming peptides. Protein Sci. 1996;5:1301–1315. doi: 10.1002/pro.5560050709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mayo KH, Haseman J, Ilyina E, Gray B. Designed beta-sheet-forming peptide 33mers with potent human bactericidal/permeability increasing protein-like bactericidal and endotoxin neutralizing activities. Biochim Biophys Acta. 1998;1425:81–92. doi: 10.1016/s0304-4165(98)00053-1. [DOI] [PubMed] [Google Scholar]
- 15.Griffioen AW, van der Schaft DW, Barendsz-Janson AF, Cox A, Struijker Boudier HA, Hillen HF, Mayo KH. Anginex, a designed peptide that inhibits angiogenesis. Biochem J. 2001;354:233–242. doi: 10.1042/0264-6021:3540233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayo KH, Griffioen AW. Designing βpep peptides: a rational approach to the discovery of novel pharmaceutical agents and small molecules. Drugs of the Future. 2003;28:337–346. [Google Scholar]
- 17.Dings RPM, Yokoyama Y, Ramakrishnan S, Griffioen AW, Mayo KH. The designed angiostatic peptide anginex synergistically improves chemotherapy and anti-angiogenesis therapy with angiostatin. Cancer Res. 2003;63:382–385. [PubMed] [Google Scholar]
- 18.Dings RPM, Hargittai B, Haseman J, Griffioen AW, Mayo KH. Anti-tumor activity of the novel angiogenesis inhibitor anginex. Cancer Letters. 2003;194:55–66. doi: 10.1016/s0304-3835(03)00015-6. [DOI] [PubMed] [Google Scholar]
- 19.Thijssen VLJL, Postel R, Brandwijk RJMGE, Dings RPM, Nesmelova I, Satijn S, Verhofstad, Nakabeppu Y, Baum LG, Bakkers J, Mayo KH, Poirier F, Griffioen AW. Galectin-1 is essential in tumor angiogenesis and is a target for anti-angiogenesis therapy. Proc. Natl. Acad. Sci. USA. 2006;103:15975–15980. doi: 10.1073/pnas.0603883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akerman ME, Pilch J, Peters D, Ruoslahti E. Angiostatic peptides use plasma fibronectin to home to angiogenic vasculature. Proc Natl Acad Sci USA. 2005;102:2040–2045. doi: 10.1073/pnas.0409844102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayo KH, van der Schaft DW, Griffioen AW. Designed β-sheet peptides that inhibit proliferation and induce apoptosis in endothelial cells. Angiogenesis. 2001;4:45–51. doi: 10.1023/a:1016672117477. [DOI] [PubMed] [Google Scholar]
- 22.Ilyina E, Roongta V, Mayo KH. NMR structure of a de novo designed, peptide 33mer with two distinct, compact β-sheet folds. Biochemistry. 1997;36:5245–5250. doi: 10.1021/bi963064o. [DOI] [PubMed] [Google Scholar]
- 23.Lockwood NA, Mayo KH. The future of antibiotics: bacterial membrane disintegrators. Drugs of the Future. 2003;28:911–923. [Google Scholar]
- 24.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 25.Brunger AT. X-plor Manual. Yale University Press; New Haven: 1992. [Google Scholar]
- 26.Mayo KH, Haseman J, Young HC, Mayo JW. Structure-function relationships in novel peptide dodecamers with broad-spectrum bactericidal and endotoxin-neutralizing activities. Biochem J. 2000;349:717–728. doi: 10.1042/bj3490717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14(51−5):29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 28.Wüthrich K. NMR of Proteins and Nucleic Acids. John Wiley & Sons, Inc.; New York: 1986. [Google Scholar]
- 29.Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. Academic Press; San Diego: 1996. [Google Scholar]
- 30.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 31.Mayo KH, Dings RPM, Flader C, Hargittai B, van der Schaft, van Eijk LI, Walek D, Haseman J, Hoye TR, Griffioen AW. Design of a partial-peptide mimetic of anginex with antiangiogenic and anticancer activity. J. Biol. Chem. 2003;278:45746–45752. doi: 10.1074/jbc.M308608200. [DOI] [PubMed] [Google Scholar]
- 32.Dings RPM, Chen X, Hellebrekers DMEI, van Eijk LI, Zhang Y, Hoye TR, Griffioen AW, Mayo KH. Design of nonpeptide topomimetics of antiangiogenic proteins with antitumor activities. J Natl Can Inst. 2006;98:932–936. doi: 10.1093/jnci/djj247. [DOI] [PubMed] [Google Scholar]