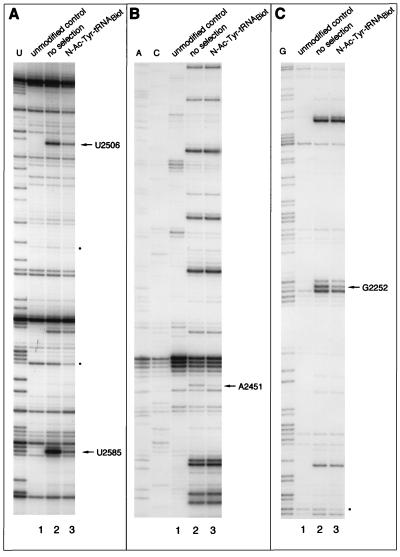
Figure 3.
Analysis of base modifications of 23S rRNA in randomly modified 50S ribosomal subunits before and after selection on the avidin resin. (A) Modification with 1-cyclohexyl-3-(2-morpholinoethyl)carbodiimide. (B) Modification with dimethyl sulfate. (C) Modification with kethoxal. rRNA was extracted from the 50S subunits, and modifications were localized by primer extension. Lanes: 1, 23S rRNA from the unmodified 50S subunits; 2, modified, unselected 50S subunits; 3, modified subunits complexed in the presence of sparsomycin with N-Ac-Tyr-tRNA and captured on the avidin resin. Dideoxy sequencing lanes corresponding to RNA bases U, A, C, and G are shown. Dots show the reverse transcriptase “strong stop” bands whose intensity is decreased in the selected samples.