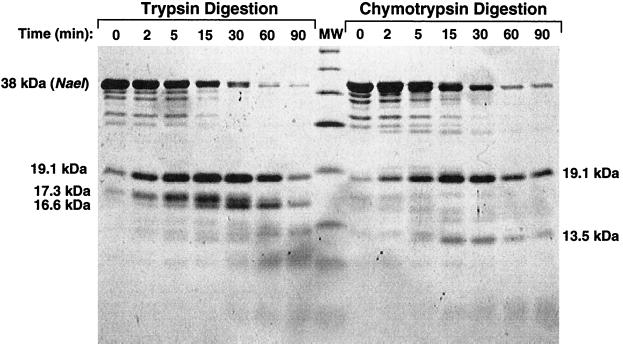
Figure 1.
Coomassie brilliant blue stained SDS-polyacrylamide gel showing pattern of polypeptide fragments produced by limited trypsin and chymotrypsin digestion of NaeI protein. The time of digestion is indicated along the tops of the lanes. The Mr of protease-resistant fragments analyzed in this study are indicated alongside the gel image and are based on the molecular weight (MW) markers glutamic dehydrogenase (55,561 Da), maltose-binding protein (42,710 Da), lactate dehydrogenase (36,487 Da), triosephosphate isomerase (26,625 Da), trypsin inhibitor (20,040–20,167 Da), lysozyme (14,313 Da), and aprotinin (6,517 Da). The digestion pattern at 0 min is caused by the amount of digestion taking place during mixing and sampling.