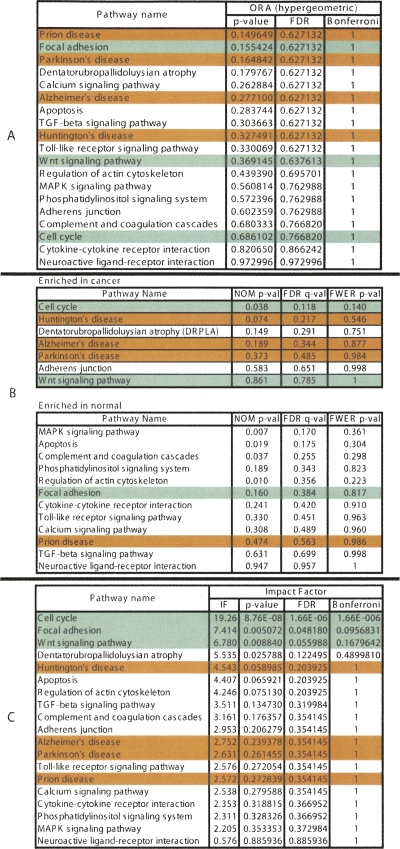
Figure 2.
A comparison between the results of the classical probabilistic approaches (A, hypergeometric; B, GSEA) and the results of the pathway impact analysis (C) for a set of genes differentially expressed in lung adenocarcinoma. The pathways marked with green are considered most likely to be linked to this condition in this experiment. The ones in red are unlikely to be related. The ranking of the pathways produced by the classical approaches is very misleading. According to the hypergeometric model, the most significant pathways in this condition are: prion disease, focal adhesion, and Parkinson’s disease. Two out of these three are likely to be incorrect. GSEA yields cell cycle as the most enriched pathway in cancer, but three out of the four subsequent pathways are clearly incorrect. In contrast, all three top pathways identified by the impact analysis are relevant to the given condition. The impact analysis is also superior from a statistical perspective. According to both hypergeometric and GSEA, no pathway is significant at the usual 1% or 5% levels on corrected P-values. In contrast, according to the impact analysis, the cell cycle is significant at 1%, and focal adhesion and Wnt signaling are significant at 5% and 10%, respectively.