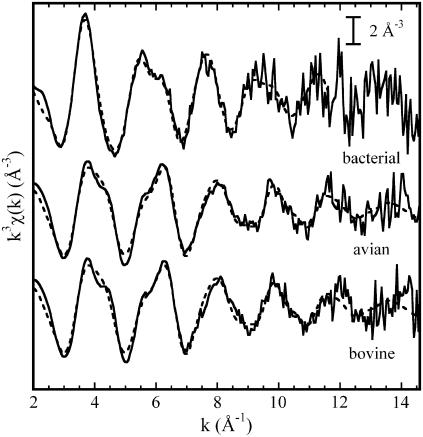
FIGURE 2.
Experimental k3 weighted EXAFS functions measured in the bacterial, avian, and bovine cyt bc1 complexes (solid lines). The dashed lines represent calculated best-fit EXAFS functions corresponding to the following clusters: two His, one Lys and one Asp or Glu residue for the avian and bovine enzymes (the corresponding structural parameters are given in Tables 2 and 3 (model m), respectively); one His, two Asp or Glu residues in monodentate coordination, one Gln or Asn residue, and two water molecules for the bacterial enzyme (see Table 4, model e, for the corresponding structural parameters).