TABLE 4.
Structural parameters determined by fitting the experimental data of the bacterial cyt bc1 complex to the model clusters extracted from the metalloprotein database MDB
| Model | Ligand cluster n | Zn-N (Å) | Zn-O (Å) | β (°) His | γ (°) Asp/Glu | η (°) Gln/Asn | R (%) | 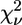 |
|---|---|---|---|---|---|---|---|---|
| a | 2 His (2 Nɛ2) | 2.05 (2) | 120 (5) | 11.0 | 8.05 (2.3) | |||
| 2 Asp/Glu m | 2.14 (3) | 123 (13) | ||||||
| 1 H2O | 2.09 (5) | |||||||
| n = 5 | ||||||||
| b | 1 His (1Nɛ2) | 2.01 (4) | 120 (11) | 9.3 | 5.4 (1.6) | |||
| 2 Asp/Glu m | 2.08 (6) | 124 (3) | ||||||
| 1 Gln/Asn | 2.18 (9) | 115 (6) | ||||||
| 1 H2O | 2.10 (17) | |||||||
| n = 5 | ||||||||
| c | 1 His (1 Nɛ2) | 2.01 (7) | 128 (40) | 14.5 | 24.3 (7.4) | |||
| 1 Asp/Glu b | 2.11 (7) | 122 (2) | ||||||
| 1 Gln/Asn | 2.10 (8) | 115 (7) | ||||||
| 1 H2O | 2.10 (17) | |||||||
| n = 5 | ||||||||
| d | 1 His (1 Nɛ2) | 2.14 (4) | 146 (14) | 10.3 | 5.7 (1.7) | |||
| 3 Asp/Glu m | 2.06 (2) | 124 (1) | ||||||
| 1 Gln/Asn | 2.22 (5) | 115 (4) | ||||||
| n = 5 | ||||||||
| e | 1 His (1Nɛ2) | 1.99 (3) | 128 (17) | 8.6 | 4.1 (1.2) | |||
| 2 Asp/Glu m | 2.10 (7) | 123 (3) | ||||||
| 1 Gln/Asn | 2.28 (4) | 111 (6) | ||||||
| 2 H2O | 2.10 (7) | |||||||
| n = 6 |
For details, go to (http://metallo.scripps.edu/advanced/#advanced_form). Monodentate and bidentate binding configurations are indicated by m and b, and n is the coordination number. Other symbols and values in parentheses are as in Table 2. See text for explanation.
