TABLE 5.
Structural parameters obtained by fitting the spectrum of the bacterial cyt bc1 complex to additional clusters containing two His residues
| Model | Ligand cluster n | Zn-N (Å) | Zn-O (Å) | β (°) His | γ (°) Asp/Glu | η (°) Gln/Asn | R (%) | 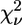 |
|---|---|---|---|---|---|---|---|---|
| a | 2 His (2 Nɛ2) | 2.05 (3) | 115 (5) | 10.6 | 7.4 (2.2) | |||
| 2 Asp/Glu m | 2.11 (2) | 124 (2) | ||||||
| 1 Gln/Asn | 2.18 (5) | 116 (4) | ||||||
| n = 5 | ||||||||
| b | 2 His (2 Nɛ2) | 2.08 (3) | 137 (7) | 15.4 | 18.4 (5.5) | |||
| 1 Asp/Glu m | 2.18 (8) | 122 (4) | ||||||
| 1 Asp/Glu b | 2.11 (7) | 121 (4) | ||||||
| n = 5 | ||||||||
| c | 2 His (2 Nɛ2) | 2.02 (3) | 118 (4) | 11.5 | 13.1 (3.8) | |||
| 2 Asp/Glu m | 2.15 (3) | 123 (1) | ||||||
| 2 H2O | 2.09 (1) | |||||||
| n = 6 | ||||||||
| d | 2 His (2 Nɛ2) | 2.05 (5) | 134 (7) | 11.8 | 12.8 (3.9) | |||
| 1 Asp/Glu m | 2.19 (6) | 123 (4) | ||||||
| 1 Asp/Glu b | 2.16 (9) | 126 (2) | ||||||
| 1 H2O | 1.93 (7) | |||||||
| n = 6 | ||||||||
| e | 2 His (2 Nɛ2) | 2.02 (3) | 135 (6) | 10.2 | 10.6 (3.3) | |||
| 1 Asp/Glu m | 2.13 (10) | 123 (11) | ||||||
| 1 Asp/Glu b | 2.13 (9) | 124 (2) | ||||||
| 1 Gln/Asn | 2.13 (10) | 116 (15) | ||||||
| n = 6 | ||||||||
| f | 2 His (2 Nɛ2) | 2.07 (3) | 135 (5) | 23.6 | 37.9 (10.8) | |||
| 2 Asp/Glu b | 2.14 (3) | 122 (2) | ||||||
| n = 6 | ||||||||
| g | 2 His (2 Nɛ2) | 2.09 (4) | 135 (5) | 17.8 | 34.2 (10.2) | |||
| 1 Asp/Glu b | 2.25 (7) | 124 (3) | ||||||
| 2H2O | 2.11 (3) | |||||||
| n = 6 | ||||||||
| h | 2 His (2 Nɛ2) | 2.06 (2) | 137 (5) | 11.7 | 13.2 (3.8) | |||
| 1 Asp/Glu m | 2.28 (5) | 118 (3) | ||||||
| 3 H2O | 2.12 (2) | |||||||
| n = 6 |
Symbols are as in Table 2. See text for details.
