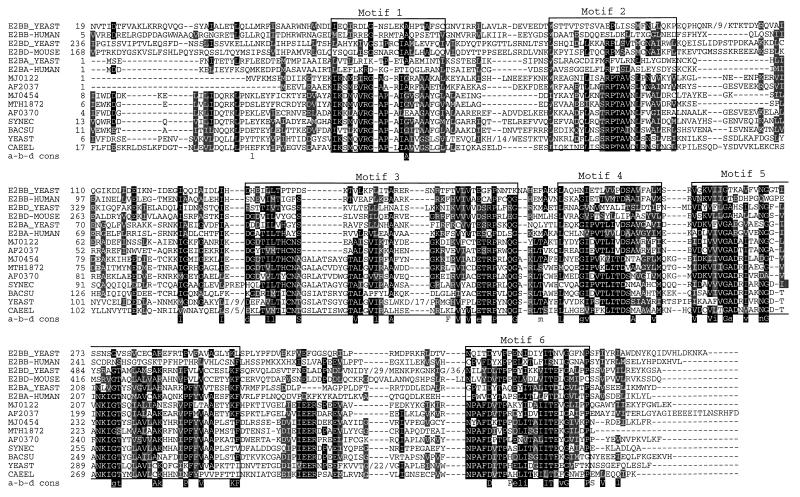
Figure 1.
Multiple sequence alignment of the eIF-2B α-β-δ subunits with their archaeal and bacterial homologs. Identical residues are highlighted black, and conservative substitutions are highlighted gray. Boxed areas mark the motif borders as generated from the meta-MEME motif search tool (29). The archaeal family of aIF-2B subunits appears to have all of the conserved residues shared between the three eukaryotic subunits, together with additional characteristics, absent from any of these three subunits. In particular, motif 1 is absent from all three eukaryotic subunits, and motifs 3, 5, and 6 are well conserved in all three of them. Motif 2 seems to be in part shared only with the δ subunit, whereas motif 4 is shared with the α and β subunits. Finally, motif 5 is less conserved in the β subunits. Protein names and their reference (accession number, when different) are: E2BB YEAST, S. cerevisiae eIF-2B β subunit; E2BB-HUMAN, human eIF-2B β subunit (YS20 HUMAN); E2BD YEAST, S. cerevisiae eIF-2B δ subunit; E2BD-MOUSE, Mus musculus eIF-2B δ subunit (PIR:B55146); E2BA YEAST, S. cerevisiae eIF-2B α subunit; E2BA-HUMAN, human eIF-2B α subunit (EMBL:X95648); MJ0122, M. jannaschii ORF MJ0122 (SW:E2B2 METJA); AF2037, A. fulgidus ORF AF2037 (gi2648498); MJ0454, M. jannaschii ORF MJ0454 (SW:E2B1 METJA); MTH1872, M. thermoautotrophicum ORF MTH1872 (gi2623009); AF0370, A. fulgidus ORF AF0370 (gi2650263); SYNEC, Synechocystis sp. hypothetical protein slr1938 (EMBL:Z99111 27); BACSU, hypothetical gene ykrs (EMBL:D90915 85); YEAST, S. cerevisiae hypothetical protein YPR118w (PIR:S69011); CAEEL, C. elegans hypothetical protein C01G10.9 (EMBL:Z81030 4). The correct annotations of the M. jannaschii functions are available at: http://geta.life.uiuc.edu/~nikos/Methanococcus.html.