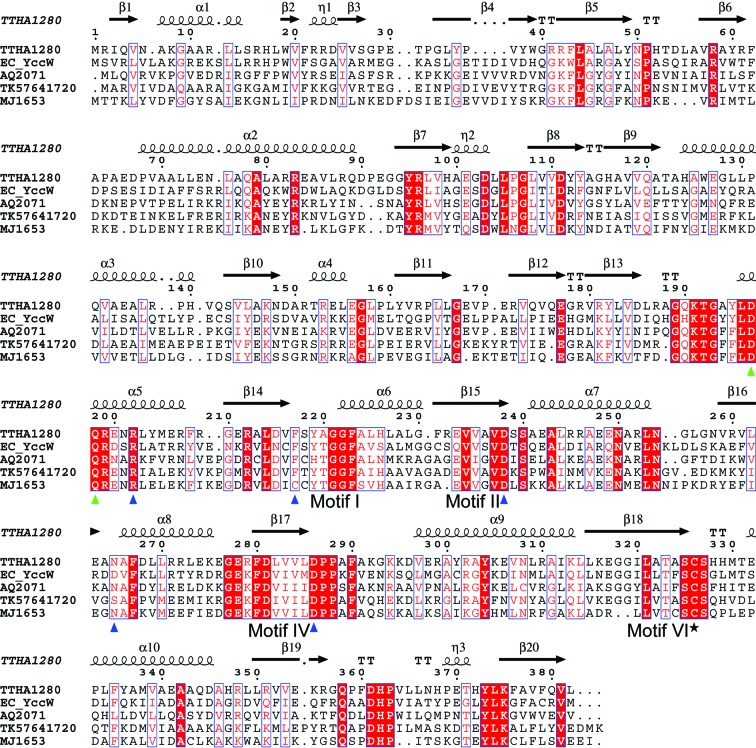
Figure 1.
Amino-acid sequence alignment of selected MJ1653 family proteins from bacterial and archaeal species. The secondary-structure elements of TTHA1280 are shown above the sequence. Identical residues are highlighted with a red background and similar residues are shown in red text with a white background. The putative catalytic cysteine residue is indicated with a black star, residues that form hydrogen bonds or hydrophobic contacts with the AdoHcy ligand are indicated with blue triangles and residues with possible roles in substrate base recognition (as described in text) are indicated with green triangles. The alignment was performed with CLUSTALW (Thompson et al., 1994 ▶) and the figure was generated with ESPript (Gouet et al., 1999 ▶). The bacterial species are Thermus thermophilus (TTHA), Escherichia coli (EC) and Aquifex aeolicus (AQ). The archaeal species are Thermococcus kodakaraensis (TK) and Methanococcus jannaschii (MJ).