Figure 6. Stereoviews of the AnsA active site.
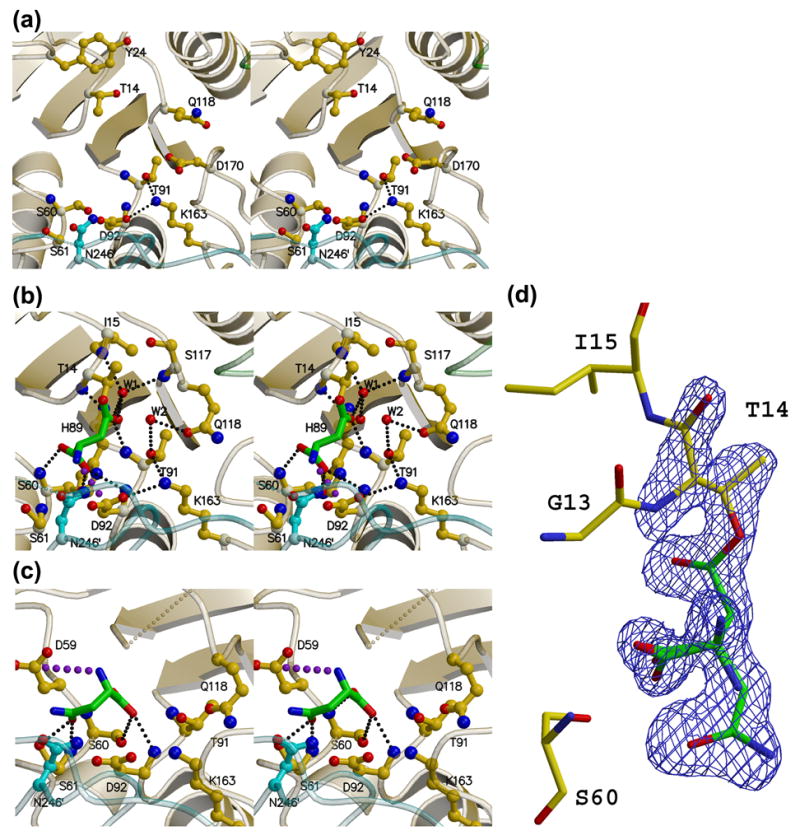
(a) The apo enzyme (APOM). (b) The AnsA-asparagine complex showing the covalently attached product aspartate (green) at the active site. Water molecule W2 hydrogen bonded to Thr91 and Gln118 is ideally positioned to act as the nucleophile that will release the product (green). (c) The AnsA-asparagine complex showing asparagine (green) bound in the non-productive alternate conformation. In each panel, monomer A is yellow, B is green and C is cyan. Details of these interactions are provided in the text, but the key active site residues are Thr14, Thr91, Lys163 and Asp92. Note how Gln118 changes conformation as the tetramer is compacted in (b) versus (a). Note also the pseudo mirror symmetry in the active site that accommodates the two alternate binding modes in (b) and (c). Large purple dots indicate a salt bridge, while small, light gray dots indicate missing structural elements. (d) Electron density observed in the active site that was interpreted as overlapped aspartate and asparagine in the non-productive alternate conformation. The Fo-Fc simulated annealing omit map is displayed at a contour level of 4σ (blue).
