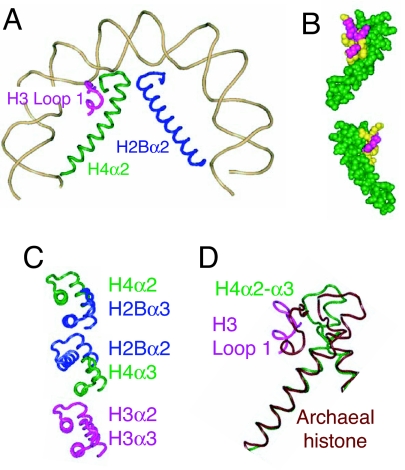
Fig. 3.
A structural change proposed to underlie the transition from wild-type to mutant Cid. (A) Half-nucleosome showing the juxtaposition of H3 loop 1 (magenta), H4α2 (green), and H2Bα2 (blue) and the bending of H4α2 around the subdomain created by H3 loop 1 + H4 loop 2 [Protein Data Bank (PDB) ID code 1KX5] (6). (B) Space-fill model showing top and bottom views of the interactions between H3 loop 1 residues whose CenH3 counterparts are critical for targeting to centromeres (yellow) and H4α2 + loop 2 (green). (C) Alignment of α2 and α3 helices comprising the three four-helix bundles that hold together conventional octameric nucleosomes. The three-dimensional structure of the nucleosome shows that H2Bα2 and H4α3 (Middle) are misaligned relative to alignments of H4α2 with H2Bα3 (Top) and H3α2 with H3α3 (Bottom) (6). It is proposed that H2Bα2 and H4α3 become well aligned in CenH3 nucleosomes, thus strengthening the four-helix bundle in the middle of the CenH3 core. (D) Aligning the dimeric histone from M. kandleri (PDB ID code 1F1E) with H3/H4 at the N-terminal residues of H4α2 [from yeast Asf1-H3/H4 (PDB ID code 2HUE] shows that the corresponding helix in this tetrameric archaeal histone is straight, whereas H3/H4 bends tightly around the H3 loop1 + H4 loop 2 subdomain.