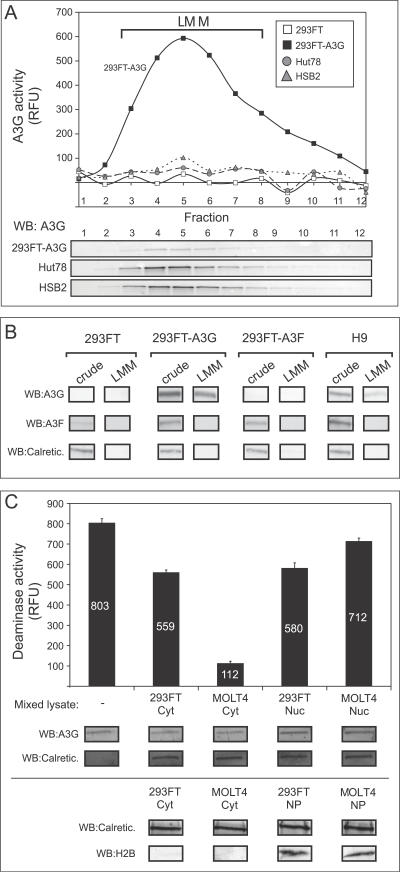
Figure 7. Active and Inhibited A3G Can Be Purified by Velocity Sedimentation.
(A) HMM A3G fractions from Hut78, HSB2, 293FT cells, or 293FT cells transfected with A3G plasmid DNA (293FT-A3G) were isolated, treated with RNase A, subjected to velocity sedimentation, and fractionated (1–12). Each fraction was assayed by both the FRET-based deaminase assay (graph) and by western blotting (WB). Migration of LMM A3G is indicated by brackets.
(B) HMM A3G from 293FT cells, 293FT cells transfected with A3G plasmid DNA, 293FT cells transfected with A3F plasmid DNA, and H9 cells were treated with RNase and subjected to velocity sedimentation and fractionated. Aliquots of crude cell lysates and pooled gradient fractions 3–8 (LMM) were analyzed by western blot (WB) with antiserum specific to A3G, A3F, or calreticulin (Calretic.), and are shown below.
(C) Cytoplasmic (Cyt) and nuclear (Nuc) extracts were prepared from either 293FT or MOLT4 T cells. Five μg of total protein from each extract, or water (-), was mixed with aliquots of pooled gradient fractions 3–8 from A3G-transfected 293FT cells containing 6 μg A3G (A and B), and A3G deaminase activity was determined using the FRET assay and was graphed. Below, western blots (WB) performed in parallel show the amount of A3G and the cellular control calreticulin (Calretic.) in each reaction. Note that the small amount of inhibition of deaminase activity seen upon addition of 293FT cytoplasmic extract and both nuclear extracts is most likely due to high levels of total protein input, which we previously observed to be mildly inhibitory (see Figure 3B). In an independent experiment (bottom), cytoplasmic extracts (Cyt) and nuclear pellets (NP) were analyzed for calreticulin (Calretic.) and the nuclear marker histone 2B (H2B). Error bars represent the standard error of the mean from triplicate reactions. Numbers on/above bars indicate actual values graphed.
(A and C) All FRET assays were performed with RNase A. Background fluorescence obtained using GGG-containing oligonucleotides was subtracted from signal detected using the CCC-containing substrate.