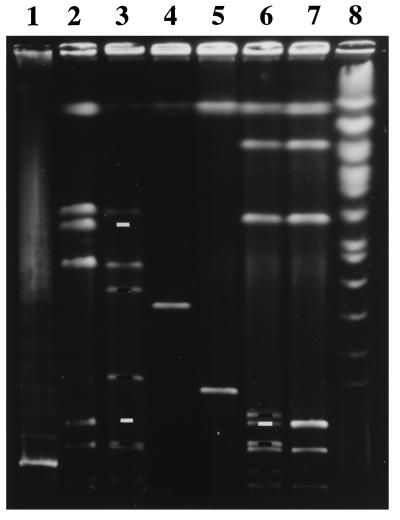
Figure 1.
Pulse-field gel electrophoresis separation of genomic segments delimited by a pair of Tn10dRCP2 insertions. Lanes: 1 and 8, yeast-chromosome and λ-concatemer standards; 2, 3, 6, and 7, I-Ceu I digests of MG1655 (parent E. coli K-12), χM2500 (K-12 double-insertion mutant), BS573 (S. flexneri 2a double-insertion mutant), and BS103 (parent S. flexneri 2a), respectively; 4 and 5, χM2500 and BS573 digested with I-Sce I. The MG1655 native I-Ceu I fragments of 130 and 670 kb were cleaved into pairs of subfragments of 30 and 100 kb and 235 and 435 kb, respectively, in the double insertion mutant χM2500 [as predicted from the NotI and Bln I map coordinates of its insertions (11) and the native I-Ceu I map (31)]. The I-Ceu I pattern of genomic DNA from strain BS573 (containing the identical two insertions in the BS103 background) showed a new subfragment of 135 kb (consistent with cleavage and lack of change in migration of the largest native I-Ceu I fragment) and a pair of subfragments of 40 and 100 kb from cleavage of one of two 140-kb native I-Ceu I fragments, consistent with P1 transduction fidelity between strains and with genetic map conservation judged by rrl gene architecture (31). Digestion of χM2500 and BS573 with I-Sce I resulted in isolated bands allowing side-by-side comparison of a pair of I-Sce I restriction fragments (398 kbMG1655 and 205 kbBS103) with corresponding end points from the MG1655 and BS103 backgrounds. White bars indicate the wild-type I-Ceu I fragments missing because of Tn10dRCP2 insertion, and black bars indicate the corresponding I-Ceu I subfragments generated.