Figure 1.
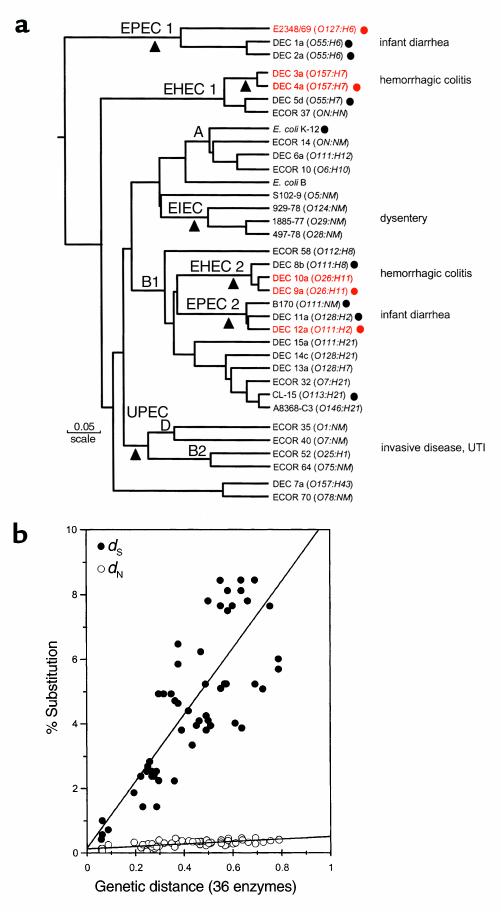
(a) The dendrogram is based on analysis of polymorphism at 36 protein loci studied by multilocus enzyme electrophoresis. Isolates mentioned repeatedly in the text are shown in red. The number of differences between strains is converted to a genetic distance assuming that each difference results from at least one amino acid–altering mutation at the DNA level. The diagram can be interpreted as a hypothetical phylogeny of strains that can be tested by gathering independent data. Main branches representing pathotypes are labeled. The A, B1, B2, and D groups are the clusters from the ECOR set. The triangles mark positions at which major acquisition of virulence factors are postulated to have occurred. (b) Nucleotide substitutions for seven housekeeping genes plotted against genetic distance. Nucleotide differences were analyzed separately for synonymous sites (dS), positions in codons where point mutations do not predict amino acid replacements, and nonsynonymous sites (dN), where point mutations result in amino acid changes. The points are averages of the comparison of pairs of strains (marked with circles) in a. UTI, urinary tract infection.
