Figure 3.
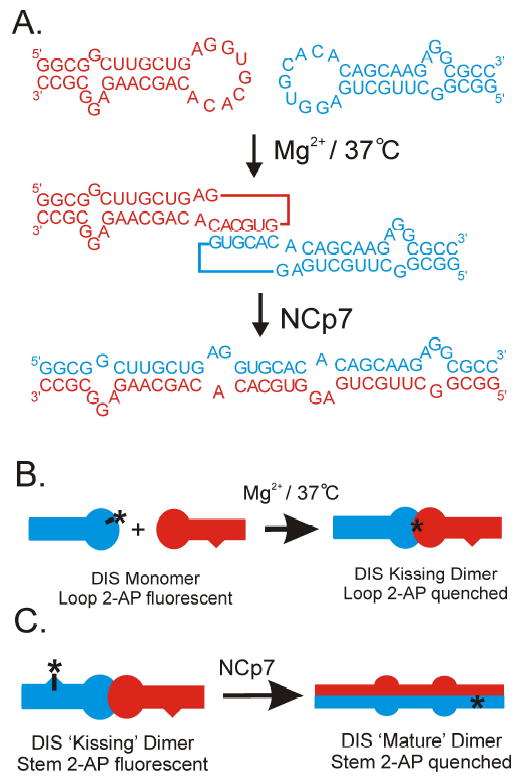
(A) Schematic of the intermolecular base pair interactions which form the basis for the DIS homodimeric loop-loop kissing complex and the structural conversion of the DIS homodimer from loop-loop kissing to extended duplex conformations. Note that one of the two identical hairpin sequences is shown in red and the other in blue. Formation of the DIS loop-loop kissing homodimer is salt dependent, with complexes formed at physiological temperature (37 °C) with divalent metals, like Mg2+, found to be kinetically trapped. At 37 °C, the conversion of Mg2+ stabilized DIS homodimer from kissing loop to extended duplex conformations requires the HIV-1 protein NCp7, which functions as a nucleic acid chaperone in stimulating and facilitating the RNA structural rearrangement. (B) Incorporation of a 2-AP, which is indicated schematically by an asterisk, within the complementary loop sequence of the DIS stem-loop allows the kissing interaction to be specifically monitored as a function of quenching of the fluorescence of the 2-AP base as it forms a stacked, base pair in the loop-loop helix. (C) Similarly, the stem strand exchange associated with conversion of the DIS stem-loop from kissing to extended duplex dimer can be monitored as a function of the quenching of a highly fluorescent bulged 2-AP again shown as an asterisk in the DIS kissing dimer that is engineered to forms a stacked, base pair in the intermolecular stem helix of the extended duplex dimer.
