Fig. 1.
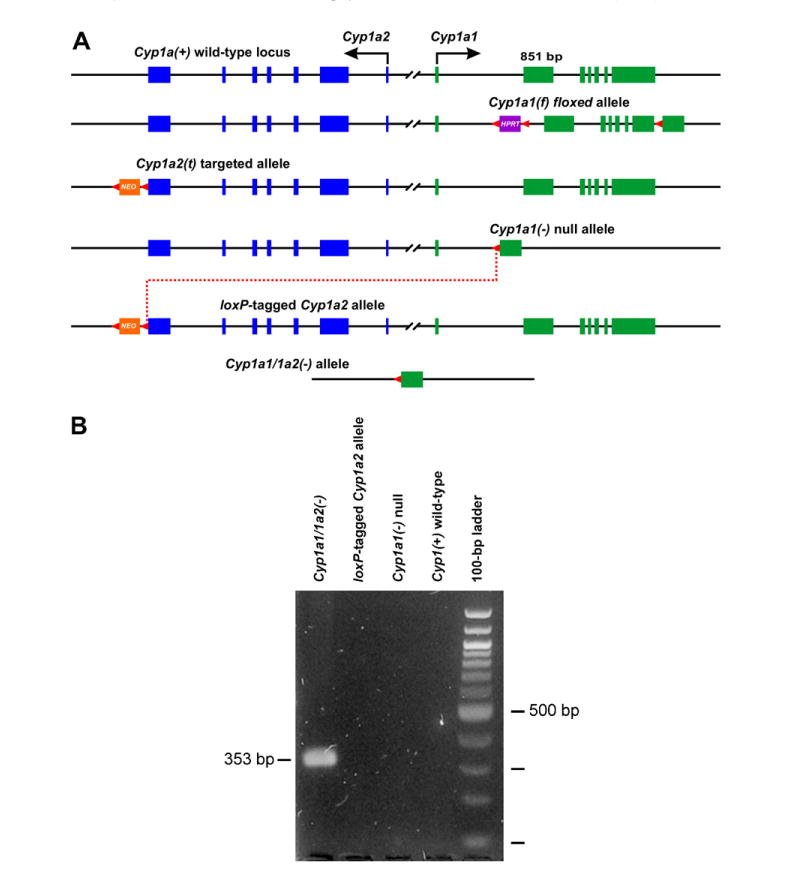
(A) Generation of the floxed Cyp1a1(f) allele, the targeted Cyp1a2(t) allele, and the Cre-mediated interchromosomal excision (dotted red line) between the Cyp1a1(−/−) null allele and the loxP-tagged Cyp1a2 allele. Cyp1a2 exons are shown in blue; Cyp1a1 exons are shown in green. All exons and introns are drawn to scale (Cyp1a1 exon 2 is 851 bp). All loxP sites (and their orientation) are shown as red arrowheads. HPRT, hypoxanthine-guanine phosphoribosyltransferase minigene used as selection marker. NEO, phosphoglycerate kinase-driven neomycin (PGK-NEO) minigene used as selection marker. For reason of clarity, most of the 13,954 bp 5′-flanking spacer region shared by both genes is omitted. (B) Confirmation of the Cyp1a1/1a2(−) allele: as expected, a 353-bp PCR fragment is generated (lane 1), compared with no such fragment for the loxP-tagged Cyp1a2 allele, the Cyp1a1(−) null allele, or the wild-type allele (lanes 2–4). The 100-bp ladder of markers is shown in far right lane. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
