Figure 6. A Promoter-Specific HMT/HDM “Code” for Regulated Transcription Units.
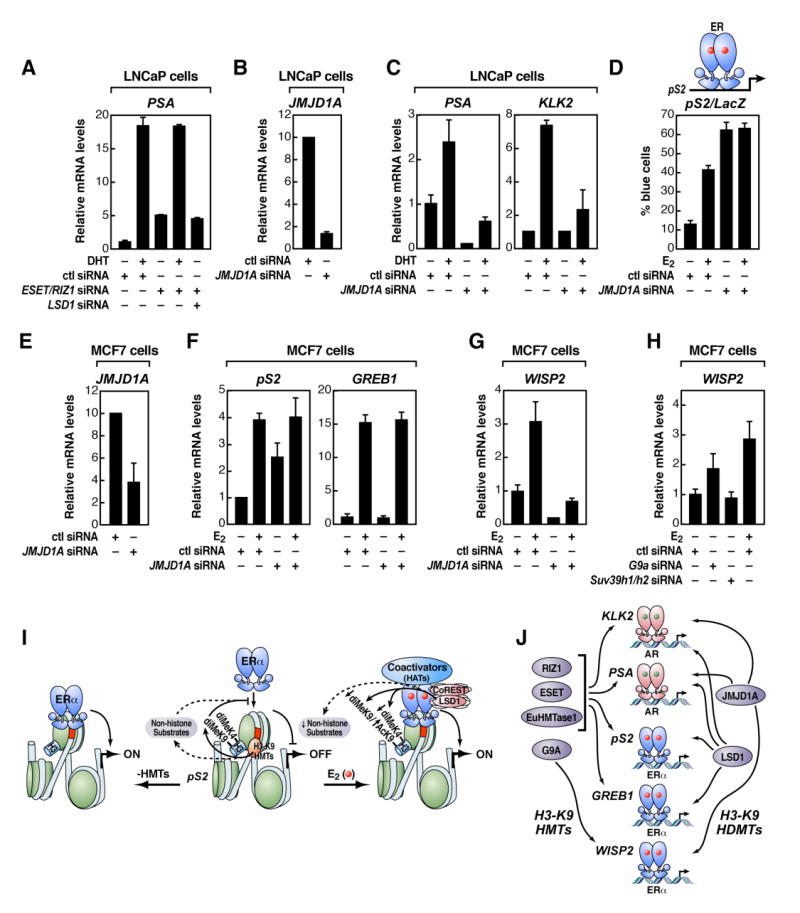
(A) RT-qPCR analysis of an endogenous LSD1+/AR+ gene target upon RIZ1/ESET and/or LSD1 depletion by siRNA in LNCaP cells is shown. (B) RT-qPCR analysis to document the efficiency of JMJD1A siRNA to diminish endogenous JMJD1A in LNCaP cells is shown. (C) RT-qPCR analysis of endogenous AR-target genes upon JMJD1A depletion by siRNA is shown. (D) Functional analysis of the pS2 promoter-LacZ reporter gene activity after removing JMJD1A by microinjection of siRNA in MCF7 cells is shown. (E) RT-qPCR analysis was performed to document efficiency of JMJD1A siRNA to diminish endogenous JMJD1A in MCF7 cells. (F) RT-qPCR gene expression analysis of endogenous LSD1+/ ERα+-target genes upon JMJD1A depletion by siRNA is shown. (G) RT-qPCR analysis of an endogenous LSD1−/ ERα+-target gene upon JMJD1A depletion by siRNA is shown. (H) RT-qPCR gene expression analysis of an endogenous LSD1−/ ERα+-target gene upon G9a or Suv39h1/h2 depletion by siRNA is shown. (I) Model of H3-K9 HMT requirement to inhibit constitutive ERα activation by blocking binding of the unliganded nuclear receptor to its cognate DNA sites is shown; HDMs, as LSD1, are required to demethylate H3-K9 HMTs substrates to permit activation by liganded ERα (see text for details). (J) Gene-specific use of HMT/HDMs to define regulated gene activation programs (see text for details) is shown. In all these experiments, siRNA was delivered by transient transfection in LNCaP (A)-(C) or MCF7 cells (D)-(H), and β-actin expression levels and cell transfection efficiency were used for normalization. The data in (A)-(H) are the average of three replicates ± standard error of the mean.
