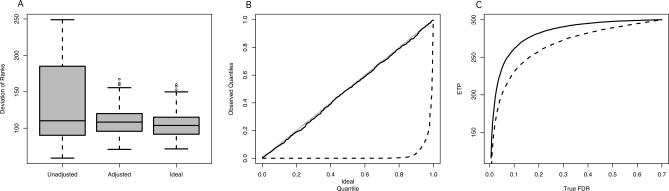
Figure 1. Impact of Expression Heterogeneity.
One thousand gene expression datasets containing EH were simulated, tested, and ranked for differential expression as detailed in Simulated Examples.
(A) A boxplot of the standard deviation of the ranks of each gene for differential expression over repeated simulated studies. Results are shown for analyses that ignore expression heterogeneity (Unadjusted), take expression heterogeneity into account by SVA (Adjusted), and for simulated data unaffected by expression heterogeneity (Ideal).
(B) For each simulated dataset, a Kolmogorov-Smirnov test was employed to assess whether the p-values of null genes followed the correct null Uniform distribution (Text S1). A quantile–quantile plot of the 1,000 Kolmogorov-Smirnov p-values are shown for the SVA-adjusted analysis (solid line) and the unadjusted analysis (dashed line). It can be seen that the SVA-adjusted analysis provides correctly distributed null p-values, whereas the unadjusted analysis does not due to EH.
(C) A plot of expected true positives versus FDR for the SVA-adjusted (solid) and -unadjusted (dashed) analyses. The SVA-adjusted analysis shows increased power to detect true differential expression.