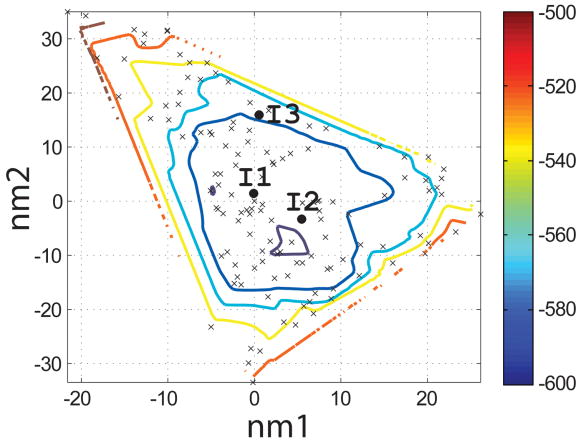
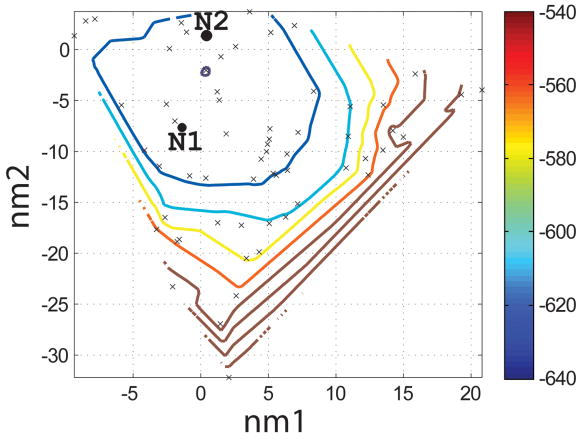
Figure 4.
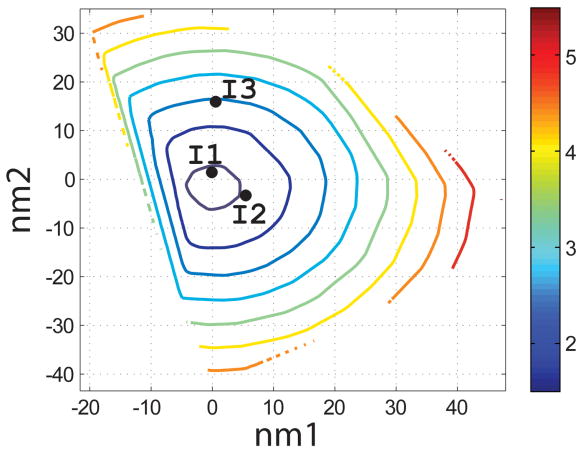
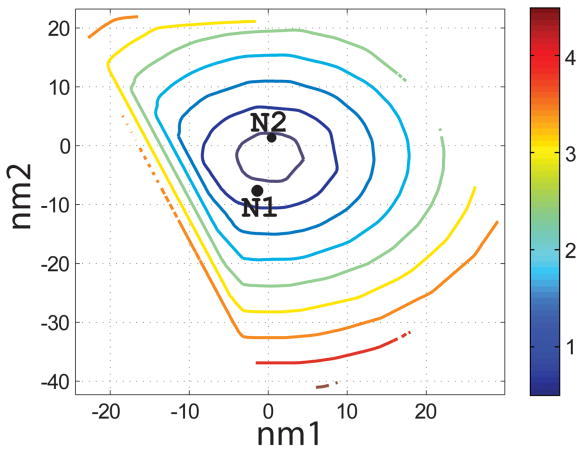
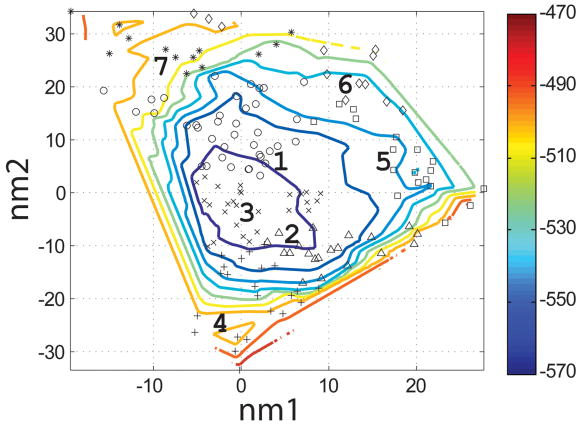
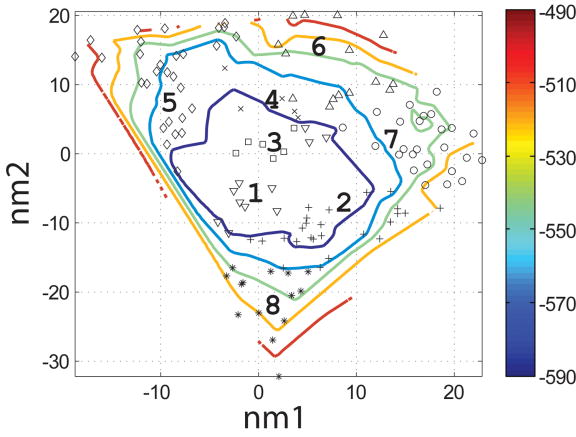
Characterization of the I- and N- sets using SCADS protein design. (a) and (b) Contour diagrams of the root-mean square deviation (rmsd) between the native backbone and (A) I-set backbones or (b) N-set backbones. (c) and (d) The Econf energy landscape of the full-sequence design of (c) I-set or (D) N-set backbones. (e) and (f) The Econf energy landscape of the design of the binding interface of (e) I-set or (f) N-set backbones. In figures (c) – (f), each symbol represents a structure for which a sequence profile was computed. In (c) and (d), structures were grouped into clusters based on the similarity of their sequence profiles from SCADS. Clusters are indicated by distinct symbols and are numbered in order of increasing energy. Color in (a) and (b) represents rmsd (Å) from the native backbone, and that in (c) – (f) represents the Econf energy (kcal/mol) interpolated from the points shown in the figures using Matlab.83 The labeled black dots in (a), (b), (e) and (f) are structures that were selected for experimental testing.