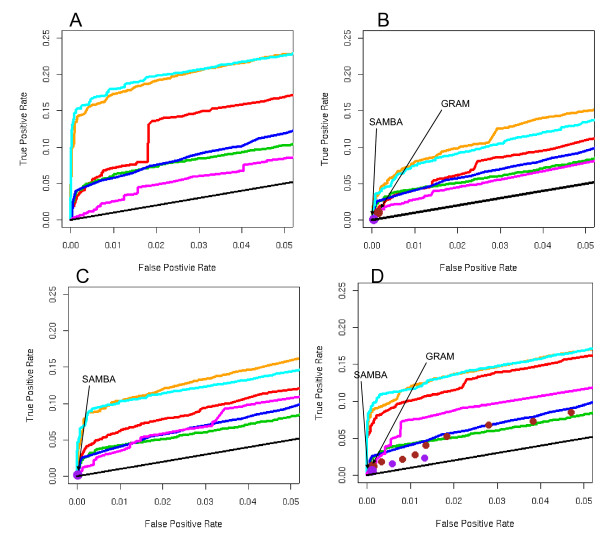
Figure 3.
ROC curves from Figures 1 and 2 with expanded range for FPR. ROC curves for 8 different algorithms using four different yeast gene expression datasets. A) Chu,1998, sporulation; B) Primig,2000, sporulation;C) Cho,1998, cellCycle, D) combined sporulation and cell-cycle dataset Liu,2006, and the ChIP-chip data of Lee, 2002. KEGG pathways were used as the gold standard. ECIM utilizing both expression and binding data dominated all other algorithms. Large ROC "spots" for GRAM and SAMBA algorithms were obtained by applying the algorithms using the default parameters. Smaller ROC "spots" for GRAM and SAMBA were obtained by systematically manipulating algorithm's parameters.