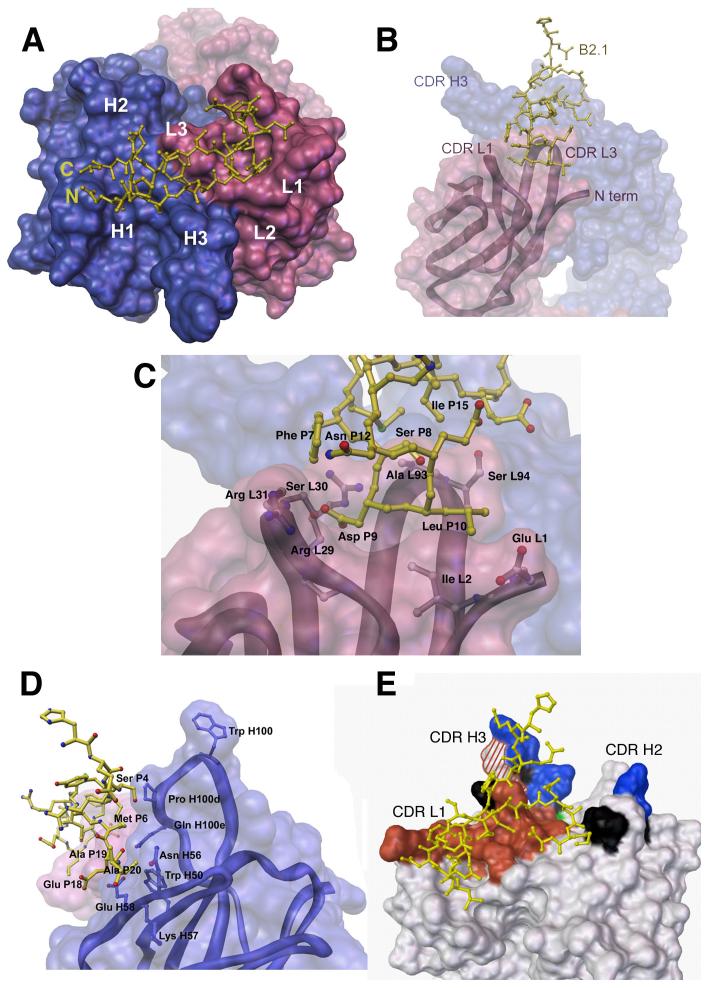
Figure 3.
B2.1-b12 interactions. In all panels, only one chain (P) of the B2.1 peptide is shown bound to its respective Fab b12 (LH), with the light chain colored pink and the heavy chain blue in panels a-d. (a) Top view of the Fab b12 antigen-binding site with the bound B2.1 peptide chain illustrated in yellow ball-and-stick representation with the N and C termini of the chain indicated. The b12 combining site is illustrated as a molecular surface with the locations of the six CDRs (L1, L2, L3, H1, H2, and H3) indicated. (b)Side view of the b12 antibody-combining site. The B2.1 chain (P) dips into a canyon formed between CDRs L1, L3 and H3. (c) Specific contacts between B2.1 chain (P) and the Fab light chain (L) involve the residues Ser 8, Asp 9 and Leu 10 of the originally-selected, SDLX3CI consensus motif. Main chain carbonyls are not pictured for clarity. (d) Side view of the b12 antibody-combining site illustrating interactions of the B2.1 chain (P) (yellow) with b12 heavy chain (H) residues Pro H100d, Gln H100e, Trp 50, Asn H56, Lys 57, and Glu H58. Heavy chain contact residues are illustrated in blue in ball and stick representation. (e) Molecular surface representation of b12 showing the antibody areas important for binding to B2.1 and b12. Shaded areas indicate substitutions that: (i) decrease binding to both B2.1 and gp120, red; (ii) have opposite effects on B2.1 and gp120 binding, red and white striped; (iii) affect gp120 binding only, blue; (iv) affect B2.1 binding only, green; and (v) have no effect on either antigen, black. Mutagenesis data are from Zwick et al. (11).