Figure 9.
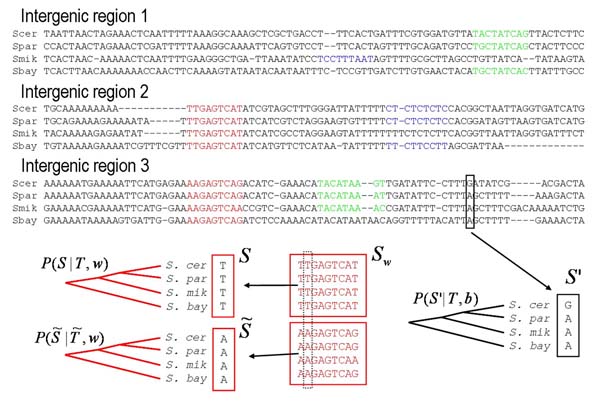
An input data-set consisting of the multiple alignments of 3 sets of orthologous intergenic regions from S. cerevisiae, S. paradoxus, S. mikatae, and S. bayanus. A binding site configuration c with sites for three motifs (red, green, and blue) is indicated. Note that each site is extended over all sequences that are locally gaplessly aligned. Most columns in the data are scored according to the background model in this configuration. On the lower right one example of an aligment column S' that is scored according to the background is shown. On the lower left the alignment Sw of sequences assigned to the red motif w is shown. A single column from this alignment consists of two independent columns, S and , that derive from the multiple alignments of intergenic regions 2 and 3 respectively. The trees on the left show that under this configuration, the columns S and are both assumed to have evolved according to the same WM w, as indicated by the red branches on their phylogenetic trees T and .
