Figure 2.
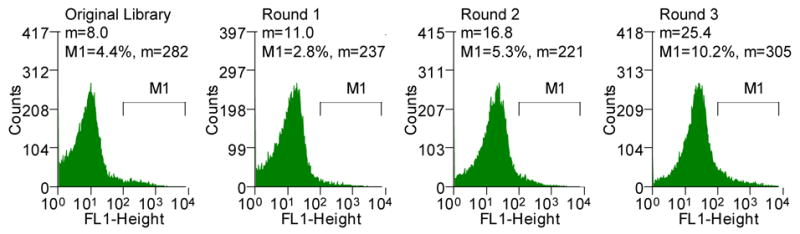
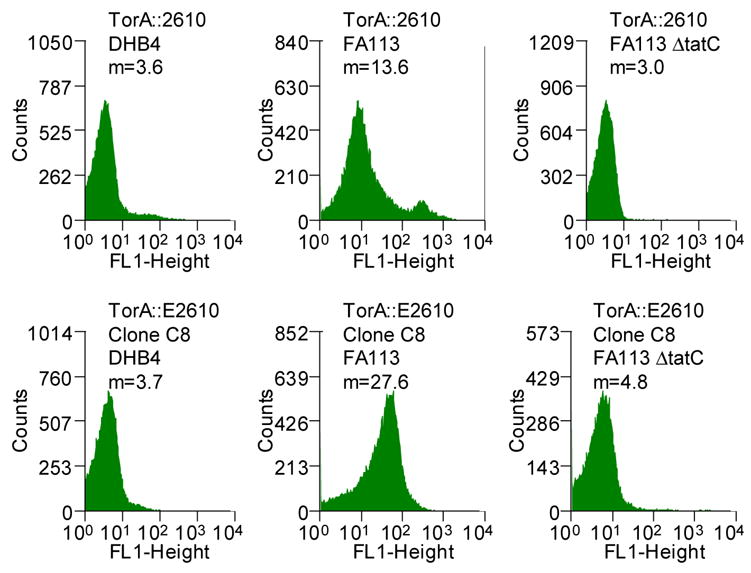
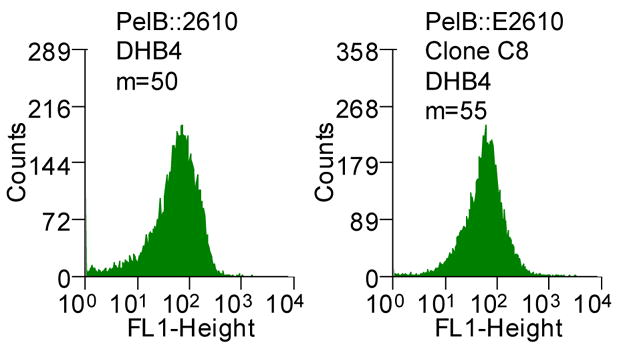
(A) Histograms showing the mean fluorescent intensities of the scFv library and the population of collected cells after each round of sorting (M1, marker indicating the percent of the total population in that region with the corresponding mean fluorescent intensity, m). The torA-2610 scFv fusion was cloned using overlap PCR and primers 1–6 (See Table S1, supplemental information). Primers 3 and 4 changed a NheI restriction site after the torA signal to an XbaI restriction site; primers 5 and 6 removed an XbaI restriction site internal to 26-10 scFv gene with a silent mutation. pTrc99AtorA-2610 (lab collection) was used as the template DNA. The amplified fragment was inserted into pBAD18(Cm). Primers 9 and 10 were used for error-prone PCR35 of the 26-10 scFv gene to create a library with approximately 13 mutations in 26-10 scFv or a 1.3% error rate which was determined by sequencing six clones from the library. The fragments were inserted into the pBAD18(Cm)torA-2610 plasmid using the XbaI and XhoI restriction sites. The ligation mixture was electroporated into the XL1-Blue (Stratagene) strain and plated for growth overnight. Plasmid was isolated from the resulting colonies by QIAprep Spin Miniprep Kit (QIAGEN). The isolated plasmids were then used to transform the library into the E. coli FA113. The library was then sorted on a MoFlo flow cytometer (FC) equipped with an Innova 90C argon laser (Cytomation, Fort Collins, CO). Triggering on side scatter was set to 488 nm light from the argon laser. Detection of digoxigenin-BODIPY binding was monitored by 518 nm emission via a 530/40 band-pass filter (FL-1). By gating for cells showing high FL-1 signal, the library of scFvs were subjected to three rounds of sorting. Between each round of sorting, the collected events were subject to colony PCR using primers 9 and 10 and reinserted into the pBAD18(Cm)torA-2610 construct using the XbaI and XhoI restriction sites. Clones isolated after round 3 PCR and transformation were picked to 96-well plates and checked by flow cytometry. (B) Fluorescent histograms of cells expressing ssTorA-C8 or the ssTorA-26-10 scFv. (C) Fluorescence histograms of E. coli DHB4 expressing ssPelB-C8 or ssPelB-26-10 scFv which are exported via Sec. The PelB signal peptide was cloned onto the pBAD18(cm)torA-2610 construct using the XmaI and XbaI restriction sites.
