Figure 1.
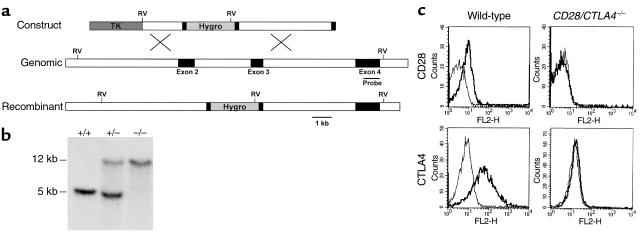
Generation of mice lacking both CD28 and CTLA4. (a) Structure of the CD28 targeting construct. The hygromycin-resistance gene is flanked by small parts of exon 2 and 3. The thymidine kinase gene (TK) lies external to the 5′ genomic fragment. The intron-exon organization of the genomic clone is shown. Exons coding for the extracellular, transmembrane, and intracellular domain of CD28 are depicted as filled rectangles. The 0.9-kb cDNA probe used for Southern blot analysis is indicated as a thin bar and is located in exon 4, external to the genomic DNA fragment used in the targeting construct. Gene targeting in ES cells by homologous recombination results in the replacement of part of exon 2, the intron between exon 2 and 3, and part of exon 3 by the hygromycin gene. Southern blot analysis uses the acquisition of an additional EcoRV site in the hygromycin gene (shown in bold) to distinguish genomic and targeted DNA. (b) Southern blot analysis of the CD28 locus. Tail DNA from wild-type (+/+), heterozygotes (+/–), and CD28/CTLA4 double-knockout (–/–) littermates were digested with EcoRV and hybridized with the probe indicated in Figure 1a, demonstrating the two bands indicative of the endogenous (12 kb) or targeted (5 kb) configuration. (c) Analysis of CD28 and CTLA4 expression. Cell-surface staining of CD28 is assayed on naive CD4 T cells; CTLA4 is stained intracellularly in activated CD4 cells. Thick lines depict staining with anti-CD28 or anti–CTLA4; thin lines depict staining with the isotype-matched hamster IgG control Ab.