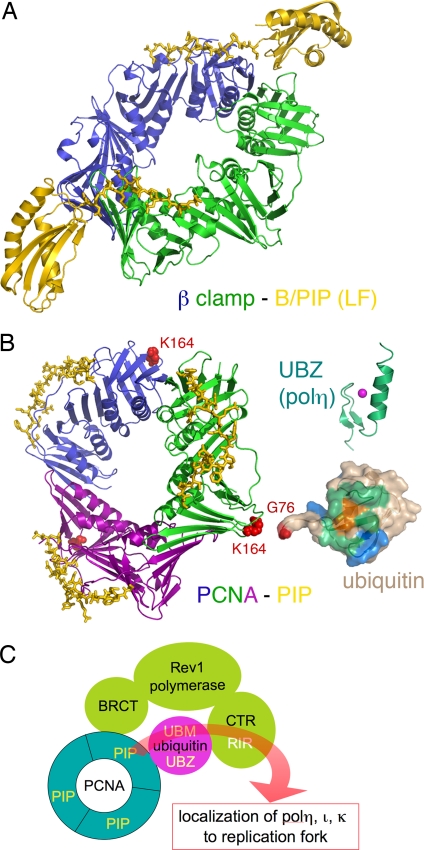
Fig. 5.
Access of Y-family polymerases to a replication fork is regulated by posttranslational modification and protein–protein interactions. (A) Ribbon diagram of E. coli polIV C-terminal region (including the LF domain and B/PIP) complexed with the β-clamp (PDB ID code 1UNN). The two subunits of β clamp are shown in green and blue, and polIV is shown in yellow. The B/PIP of polIV is represented by a stick model. (B) Interactions among PCNA–PIP (represented by p21, PDB ID code 1AXC), ubiquitin, UBZ, and UBM. The trimeric PCNA is shown in blue, green, and purple ribbon diagram. The PIP peptide from p21 is shown as yellow sticks. When ubiquitinated, PCNA is covalently linked through its K164 (represented by red spheres) with G76 (highlighted in red) of ubiquitin (PDB ID code 2G45). Ubiquitin is shown in molecular surface representation, the conserved I44 is highlighted in orange, and the surrounding areas that have been mapped to interact with UBZ and UBM are highlighted in green and blue, respectively. The NMR structure of the polη UBZ (PDB ID code 2I5O) is shown in green ribbon diagrams, and a magenta sphere represents the zinc ion. (C) A cartoon summarizing the protein–protein interactions of eukaryotic Y-family polymerases. Rev1 (pea green), polη, polι, and polκ each interact with PCNA (cyan) and ubiquitin (magenta), and the C-terminal region of Rev1 interacts with polη, polι, and polκ (collectively represented by the curvy red arrow). The multilayered interactions occur in response to DNA damage and may allow polη, polι, and polκ to be recruited to replication forks.