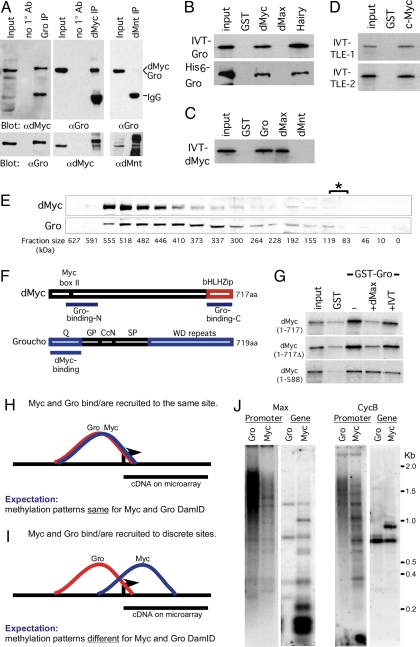
Fig. 4.
dMyc and Gro form a complex in vivo and in vitro. (A) Immunoprecipitation of dMyc–Gro complexes from stage 8 and 9 Drosophila embryos. Embryo extracts were immunoprecipitated with the indicated antibodies and analyzed by Western blot. Note that the dMyc and Gro proteins are 717 and 719 aa, respectively. (B) IVT-35S-Met-labeled full-length Gro or purified full-length His6-Gro interacts with GST-Myc (81–504 aa) and GST-Hairy. (C) IVT-35S-dMyc interacts with GST-Gro and GST-dMax. (D) Full-length IVT-35S-Met-labeled mammalian TLE-1 and -2 proteins bind to GST-c-Myc (1–262 aa). (E) dMyc and Gro protein colocalize to a ≈520-kDa fraction upon gel filtration. The asterisk denotes the fractions (83–110 kDa) where dMyc and Gro monomers would be expected to localize. (F) Diagram depicting the regions mediating dMyc–Gro interaction. (G) IVT-dMax outcompetes binding of full-length 35S-dMyc to GST-Gro (at the top). This displacement requires dMyc's bHLHZ domain. For all cases, 5% input is shown. (H–J) dMyc and Gro generate differential Dam-methylation patterns within a shared target gene locus and are likely recruited to independent sites at their shared targets. (H and I) Graphic representation of a theoretical binding and methylation pattern generated by DamID fragments from Dam-dMyc and Dam-Gro if they bind or are recruited to a single site (H) or discrete sites (I) within their shared target locus. (J) Southern blot analysis of dMyc and Gro DamID generated fragments hybridized with probes to the promoter (“promoter”) or coding regions (“gene”) of two representative dMyc-Gro shared targets, CycB and dMax (see also SI Text).