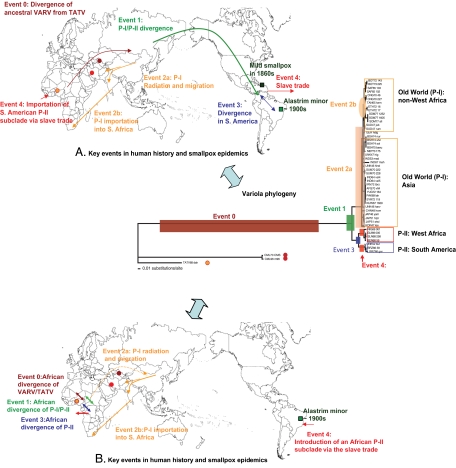
Fig. 3.
Hypotheses of the origin of VARV. Event 0, The high similarity of VARV to CMLV and TATV genome sequences suggests that TATV and CMLV share a more recent common ancestor with VARV than do other known Orthopoxvirus species. Event 1, hypothesis A: An ancestral VARV diverged into two primary clades (P-I and P-II) and evolved independently in the Old World and the New World by following its human hosts. Event 1, hypothesis B: The divergence of P-I/P-II in Africa. Event 2, hypotheses A and B: The long single branch and subsequent radiation of P-I suggest that the ancestral VARV of P-I probably originated in Northeast Asia and that the smallpox endemics of P-I started to spread to East Asia, the Middle East, and India as local populations became sufficiently large. Event 2a, The diversification and migration of P-I throughout Asia. Event 2b, Western exploration brought VARV major to South Africa from the Indian subcontinent, which then spread northward to Central Africa and the Horn of Africa (Somali Peninsula). Events 3 and 4, hypothesis A: The diversification of P-II in the New World followed by its reintroduction into Africa; hypothesis B: The diversification of P-II in Africa, followed by the later introduction of one subclade into the New World.