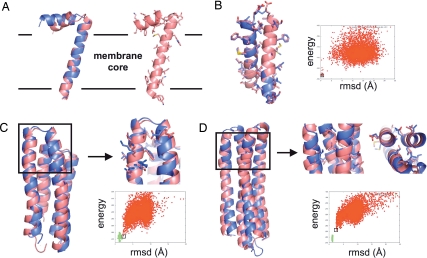
Fig. 2.
Structure prediction. (A) fd-coat protein. (Left) Backbone superposition of the experimental structure determined by solid-state NMR (19) (blue) and the lowest-energy decoy generated by ROSETTA (pink) starting from an extended chain. The rmsd over 30 Cα atoms is 2.4 Å. (Right) All-atom representation of the lowest-energy decoy generated by ROSETTA. The boundaries predicted by ROSETTA between the hydrophobic core and the interface regions of the membrane are represented with a black solid line. (B) Glycophorin A. Isolated monomers were docked with the ROSETTA protein–protein docking protocol and the all-atom membrane force field. The superposition between the native (blue) and the lowest-energy predicted structure (pink) is represented. The rmsd over 45 Cα atoms is 0.65 Å. (C and D) Ab initio structure prediction of polytopic membrane proteins. Native polytopic membrane protein conformations define a narrow energy basin in the all-atom conformational energy landscape. When near-native topologies are generated at the coarse-grained level, all-atom relaxed decoys define a funnel toward the native basin and the lowest-energy predicted structures have near-atomic resolution structures. In the energy versus rmsd plots, nonnative (red points) (generated from sequence by the ROSETTA coarse-grained structure prediction mode) and native conformations (green points) were relaxed by sampling the conformational degrees of freedom of all backbone and side-chain atoms. Cartoons show superposition between the native (blue) and the lowest-energy predicted structure (pink). Boxed areas show regions where close to native side-chain packing arrangements were obtained. The rmsd values are 2.1 Å over 111 Cα atoms for BRD4 (C) and 2.4 Å over 139 Cα atoms for VATP (D).