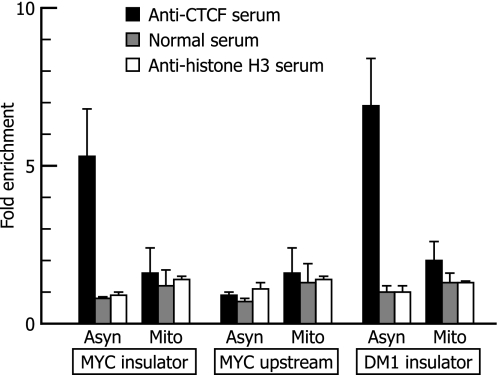
Fig. 4.
ChIP analysis of the association of CTCF with the c-MYC gene and the DM1 locus in asynchronous and mitotic cells. Values are expressed as “fold enrichment” over the input control. Chromatin from asynchronous (Asyn) and mitotic (Mito) HeLa cells was precipitated with anti-CTCF serum, normal serum, or anti-histone H3 serum. DNA was recovered from immunoprecipitated and nonimmunoprecipitated (input) chromatin and analyzed by real-time PCR. Primer pairs used for PCR were specific to the insulator or upstream region of the c-MYC gene (see Fig. 1A) or the insulator region of the DM1 locus. The fold enrichment of the target sequence in precipitated DNA compared with input DNA was calculated by comparison of the threshold cycle value of the sample of precipitated DNA with the standard curve generated from the threshold cycle values for input DNA. The means and the SD of data from three independent experiments with separate chromatin preparations are shown.