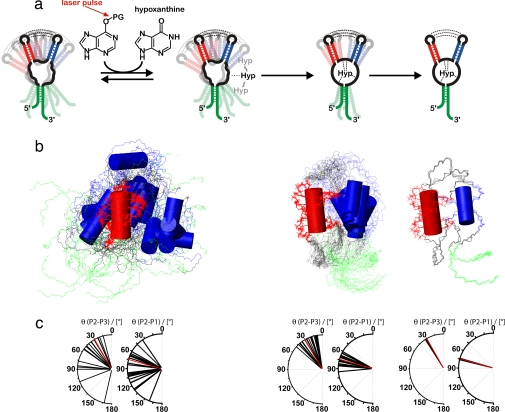
Fig. 4.
Structural interpretation of the conformational transition. (a) Schematic illustration of the proposed folding model of GSRapt-RNA on ligand binding based on the experimentally restrained torsion angle MD simulations. The first step is low-affinity binding, the second step is the ligand-binding process, and the third step is helical tightening. (b) Overlaid structures of the three states simulated according to our NMR data, aligned on helix P2 (red). Helices P3 and P1 are blue and green, respectively. (Left) The free form of GSRapt, where only the loop–loop interaction and the canonical form of the three helices are restrained. (Center) The transition state-like form where the core is folded toward the native conformation. (Right) The native structure. (c) Distribution of helix–helix projection angles [°] between helices P2/P3 and P2/P1 as seen in the crystal structure (22) (red) and in the structural ensembles depicted in Fig. 4b (black).