Table 1.
Summary of line width values obtained for 13C,15N-labeled ligands in the presence of various RNA constructs
| RNA | Secondary structure | Ligand (13C,15N-labeled) | Mg2+, mM | Characteristics | Δσ, Hz | Line width ligand, Hz |
|---|---|---|---|---|---|---|
| No RNA | Hypoxanthine | 0.11 | 7.5 ± 1.0 | |||
| GSRapt | 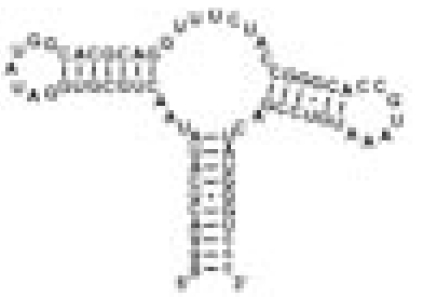 |
Hypoxanthine | Specific hypoxanthine binding | 0.13 | 31.5 ± 0.6 | |
| GSRapt | 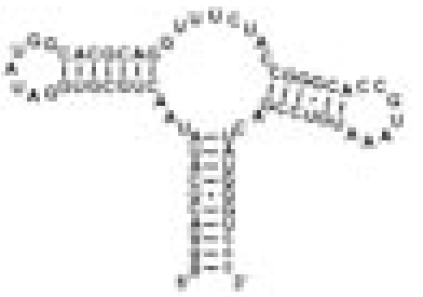 |
Hypoxanthine | 5 | Specific hypoxanthine binding | 0.12 | 30.5 ± 0.4 |
| ASRapt | 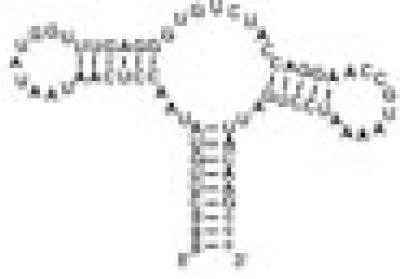 |
Hypoxanthine | No binding of hypoxanthine | 0.06 | 11.5 ± 0.5 | |
| Helix P2&P3 RNA |  |
Hypoxanthine | No binding of hypoxanthine | 0.20 | 9.6 ± 1.3 | |
| No RNA | Adenine | 0.16 | 7.8 ± 0.1 | |||
| ASRapt | 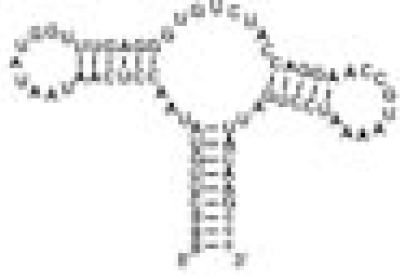 |
Adenine | 5 | Specific adenine binding | 0.04 | 27.8 ± 0.4 |
| GSRapt |  |
Adenine | No binding of adenine | 0.03 | 12.1 ± 0.1 | |
| Helix P2&P3 RNA |  |
Adenine | No binding of adenine | 0.07 | 10.3 ± 0.6 |
The RNA constructs include the guanine riboswitch aptamer domain (GSRapt), the adenine riboswitch aptamer domain (ASRapt), and an RNA construct (helix P2&P3 RNA), which consists of the structural elements helices P2 and P3 and loops L2 and L3 but lacks the ligand binding site of the purine riboswitches (see SI Fig. 5 for details) (Δσ, deviation of individual DSS reference signal from mean DSS line width value; line width ligand, mean line width value of two signals H2 and H8 of the respective ligand).
